Transcription of Eukaryotic Genes
| Home | | Biochemistry |Chapter: Biochemistry : RNA Structure, Synthesis, and Processing
The transcription of eukaryotic genes is a far more complicated process than transcription in prokaryotes.
TRANSCRIPTION OF EUKARYOTIC GENES
The transcription of
eukaryotic genes is a far more complicated process than transcription in
prokaryotes. Eukaryotic transcription involves separate polymerases for the
synthesis of rRNA, tRNA, and mRNA. In addition, a large number of proteins
called transcription factors (TFs) are involved. TFs bind to distinct sites on
the DNA either within the core promoter region, close (proximal) to it, or some
distance away (distal). They are required both for the assembly of a
transcription complex at the promoter and the determination of which genes are
to be transcribed. [Note: Each eukaryotic RNA pol has its own promoters and TFs
that bind core promoter sequences.] For TFs to recognize and bind to their
specific DNA sequences, the chromatin structure in that region must be altered
(relaxed) to allow access to the DNA. The role of transcription in the
regulation of gene expression is discussed in Chapter 32.
A. Chromatin structure and gene expression
The association of DNA
with histones to form nucleosomes affects the ability of the transcription
machinery to access the DNA to be transcribed. Most actively transcribed genes
are found in a relatively relaxed form of chromatin called euchromatin, whereas
most inactive segments of DNA are found in highly condensed heterochromatin.
[Note: The interconversion of these forms is called chromatin remodeling.] A
major component of chromatin remodeling is the covalent modification of
histones (for example, the acetylation of lysine residues at the amino terminus
of histone proteins) as shown in Figure 30.11. Acetylation, mediated by histone
acetyltransferases (HATs), eliminates the positive charge on the lysine,
thereby decreasing the interaction of the histone with the negatively charged
DNA. Removal of the acetyl group by histone deacetylases (HDACs) restores the positive
charge and fosters stronger interactions between histones and DNA. [Note: The
ATP-dependent repositioning of nucleosomes is also required to access DNA.]
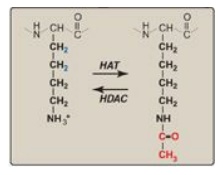
Figure 30.11 Acetylation/deacetylation of a lysine residue in a histone protein. HAT = histone acetyltransferase; HDAC = histone deacetylase.
B. Nuclear RNA polymerases of eukaryotic cells
There are three distinct classes of RNA pol in the nucleus of eukaryotic cells. All are large enzymes with multiple subunits. Each class of RNA pol recognizes particular types of genes.
1. RNA polymerase I: This enzyme synthesizes the
precursor of the 28S, 18S, and 5.8S rRNA in the nucleolus.
2. RNA polymerase II: This enzyme synthesizes the
nuclear precursors of mRNA that are subsequently translated to produce
proteins. RNA pol II also synthesizes certain small ncRNAs, such as snoRNA,
snRNA and miRNA.
a. Promoters for RNA polymerase II: In some genes transcribed by RNA
pol II, a sequence of nucleotides (TATAAA) that is nearly identical to that of
the Pribnow box is found centered about 25 nucleotides upstream of the transcription
start site. This core promoter consensus sequence is called the TATA, or Hogness,
box. In the majority of genes, however, no TATA box is present. Instead,
different core promoter elements such as Inr (initiator) or DPE (downstream
promoter element) are present (Figure 30.12). [Note: No one consensus sequence
is found in all core promoters.] Because these sequences are on the same
molecule of DNA as the gene being transcribed, they are cis-acting. The
sequences serve as binding sites for proteins known as general transcription
factors (GTFs), which in turn interact with each other and with RNA pol II.
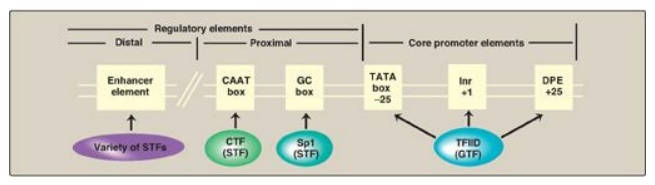
Figure 30.12 Eukaryotic gene
cis-acting promoter and regulatory elements and their trans-acting general and
specific transcription factors (GTF and STF, respectively). Inr = initiator;
DPE = downstream promoter element.
b. General transcription factors: These are the minimal requirements
for recognition of the promoter, recruitment of RNA pol II to the promoter, and
initiation of transcription at a basal level (Figure 30.13A). GTFs are encoded
by different genes, synthesized in the cytosol, and transit to their sites of
action, and so are trans-acting. [Note: In contrast to the prokaryotic
holoenzyme, eukaryotic RNA pol II does not itself recognize and bind the
promoter. Instead, TFIID, a GTF containing TATA-binding protein and
TATA-associated factors, recognizes and binds the TATA box (and other core
promoter elements). TFIIF, another GTF, brings the polymerase to the promoter.
The helicase activity of TFIIH melts the DNA, and its kinase activity
phosphorylates polymerase, allowing it to clear the promoter.]
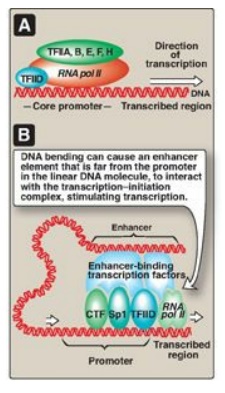
Figure 30.13 A. Association of the general transcription factors (TFIIs) and RNA polymerase II (RNA pol II) at the core promoters. [Note: The Roman numeral II denotes the TFs for RNA pol II.] B. Enhancer stimulation of transcription. CTF = CAAT box transcription factor; Sp1 = specificity factor-1.
c. Regulatory elements and transcriptional activators: Upstream of the core promoter are additional consensus sequences (see Figure 30.12). Those close to the core promoter (within 200 nucleotides) are the proximal regulatory elements, such as the CAAT and GC boxes. Those farther away are the distal regulatory elements such as enhancers. Proteins known as transcriptional activators or specific transcription factors (STFs) bind these regulatory elements. STFs bind to promoter proximal elements to regulate the frequency of transcription initiation, and to distal elements to mediate the response to signals such as hormones and regulate which genes are expressed at a given point in time. A typical protein-coding eukaryotic gene has binding sites for many such factors. [Note: STFs have two binding domains. One is a DNA-binding domain, the other is a transcription activation domain that recruits the GTFs to the core promoter as well as “coactivator” proteins such as the HAT enzymes involved in chromatin modification.]
Transcriptional activators bind DNA through a
variety of motifs, such as the helix-loop-helix, zinc finger, and leucine
zipper.
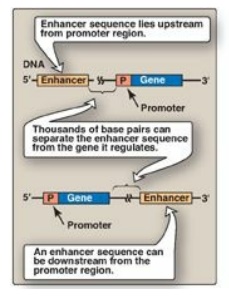
Figure 30.14 Some possible
locations of enhancer sequences.
d. Role of enhancers in eukaryotic gene regulation: Enhancers are special DNA
sequences that increase the rate of initiation of transcription by RNA pol II. Enhancers
are typically on the same chromosome as the gene whose transcription they
stimulate (Figure 30.13B). However, they can 1) be located upstream (to the 5
-side) or downstream (to the 3 -side) of the transcription start site, 2) be
close to or thousands of base pairs away from the promoter (Figure 30.14), and
3) occur on either strand of the DNA. Enhancers contain DNA sequences called
“response elements” that bind STFs (transcriptional activators). By bending or
looping the DNA, these enhancer-binding proteins can interact with other
transcription factors bound to a promoter and with RNA pol II, thereby
stimulating transcription (see Figure 30.13B). [Note: Although silencers are
similar to enhancers in that they also can act over long distances, they reduce
gene expression.]
e. Inhibitors of RNA polymerase II: α-Amanitin, a potent toxin produced
by the poisonous mushroom Amanita phalloides (sometimes called “the death
cap”), forms a tight complex with RNA pol II, thereby inhibiting mRNA
synthesis.
3. RNA polymerase III: This enzyme synthesizes tRNA, 5S
rRNA, and some snRNA and snoRNA.
C. Mitochondrial RNA polymerase
Mitochondria contain a single RNA pol that more closely resembles bacterial RNA pol than the eukaryotic nuclear enzymes.
Related Topics
