Multiple Drug Resistance
| Home | | Pharmaceutical Microbiology | | Pharmaceutical Microbiology |Chapter: Pharmaceutical Microbiology : Bacterial Resistance To Antibiotics
Several issues of multiple drug resistance have already been raised in this chapter. Notable examples are MRSA, which can harbour both small cryptic plasmids and larger plasmids encoding resistance to antiseptics, disinfectants, trimethoprim, penicillin, gentamicin, tobramycin and kanamycin, and multidrug-resistant M. tuberculosis.
MULTIPLE DRUG
RESISTANCE
R-factors
Several issues
of multiple drug
resistance have already been raised in this chapter.
Notable examples are MRSA,
which can harbour both small cryptic
plasmids and larger plasmids encoding
resistance to antiseptics, disinfectants, trimethoprim, penicillin, gentamicin, tobramycin and kanamycin, and multidrug-resistant M. tuberculosis. Of equal
concern are instances where isolates can become resistant to multiple, chemically
distinct agents in a single
biological event. One
of the earliest examples was in Japan in 1959. Previously sensitive E. coli
became resistant to multiple antibiotics through acquisition of a conjugative plasmid (R-factor) from resistant Salmonella and Shigella isolates. A number of R-factors
have now been characterized including
RP4, encoding resistance to ampicillin, kanamycin, tetracycline and neomycin, found in Ps. aeruginosa and other Gram-negative bacteria; R1, encoding
resistance to ampicillin, kanamycin, sulphonamides, chloramphenicol and streptomycin, found in Gram-negative bacteria and pSH6, encoding resistance to gentamicin, trimethoprim and
kanamycin, found in Staph. aureus.
Mobile Gene
Cassettes And Integrons
Many Gram-negative resistance genes are located in gene
cassettes. One or more of these cassettes can be integrated into a specific
position on the chromosome termed an
integron. More than 60 cassettes have been identified, each comprising only
a promotor-less single
gene (usually antibiotic resistance) and a 59-base
element forming a specific recombination site. This
recombination site confers mobility
because it is recognized by specific
recombinases encoded
by integrons that catalyse integration of the cassette into a specific
site within the integron. Thus, integrons are genetic
elements that recognize and capture multiple
mobile gene cassettes. As the gene typically lacks
a promoter, expression is dependent on correct
orientation into the integron to supply the upstream promoter. Four classes of integron have been
identified, although only
one member of class 3 has been
described and class
4 integrons are limited to Vibrio
cholerae. Analysis of the resistant Shigella strains isolated in Japan has shown that some
of the conjugative plasmids included an integron with
one or two integrated cassettes.
Chromosomal Multiple-Antibiotic Resistance (mar)
Locus
The multiple-antibiotic resistance (mar) locus
was first described in E. coli by Stuart
Levy and colleagues at Tufts University and has since been recognized in other
enteric bacteria. The locus consists
of two divergently transcribed units, marC and
marRAB. Little is known
of marC and marB;
however, marR encodes a repressor of the operon, and marA encodes a transcriptional activator affecting expression of more than 60 genes.
Increased expression of the MarRAB operon resulting from mutations in marO or
marR, or from inactivation of MarR following exposure to inducing agents such as salicylate,
leads to the Mar phenotype. This phenotype is characterized by resistance to structurally unrelated antibiotics, organic solvents, oxidative stress and chemical
disinfectants. A number
of effector mechanisms have been identified, including increased expression of the acrAB-tolC
multidrug efflux system and the soxRS regulon.
Multidrug Efflux Pumps
Whereas some efflux pumps
excrete only one drug or class of drugs,
a multidrug efflux
pump can excrete
a wide range of compounds
where there is often little
or no chemical similarity between
the substrates. One common
characteristic may be agents with a significant hydrophobic domain. For this reason, hydrophilic compounds such as the aminoglycoside antibiotics are not exported
by these
systems. A distinction needs to be drawn between those efflux systems, typically in Gram-positive bacteria, that pump their substrate
across the cytoplasmic membrane, such as the QacA and Smr pumps which
both export
quaternary ammonium compounds and basic dyes,
and those which
efflux across the cytoplasmic and outer membranes of Gram-negative bacteria. There are some examples of single
membrane systems in Gramnegative bacteria, such as the EmrE protein
in E. coli, but they
are not of great clinical
significance. The majority
of Gram-negative pumps
span both membranes and include the AcrAB-TolC system
in E. coli and
the MexABOprM system
in Ps. aeruginosa. Genomic
analyses are revealing
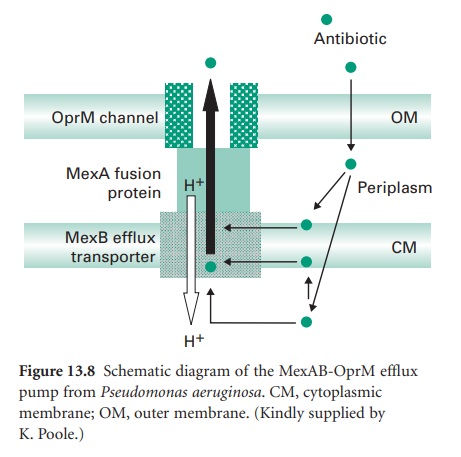
numerous homologues. Using the MexABOprM system as the prototypic example
(Figure 13.8), MexA is the linker protein
and MexB is in the cytoplasmic membrane. MexB is a resistance-nodulation-division (RND) family member
and is predicted to be a proton antiporter with 12 membrane-spanning α-helices. OprM shows homology
with outer-membrane channels
of systems
thought to export such diverse molecules as nodulation signals and alkaline proteases.
Mutations in regulatory genes such as nalB cause overexpression of MexAB-OprM and consequently multidrug resistance. MexB is a proton antiporter and efflux by this and other
members of the RND family
is energized by the proton motive force. This contrasts with
mammalian multidrug
efflux pumps (MDR) that are powered by ATP hydrolysis.
Related Topics
