DNA Repair
| Home | | Biochemistry |Chapter: Biochemistry : DNA Structure, Replication, and Repair
Despite the elaborate proofreading system employed during DNA synthesis, errors (including incorrect base-pairing or insertion of one to a few extra nucleotides) can occur.
DNA REPAIR
Despite the elaborate proofreading system employed during DNA synthesis, errors (including incorrect base-pairing or insertion of one to a few extra nucleotides) can occur. In addition, DNA is constantly being subjected to environmental insults that cause the alteration or removal of nucleotide bases. The damaging agents can be either chemicals (for example, nitrous acid, which can deaminate bases), or radiation (for example, non-ionizing ultraviolet light, which can fuse two pyrimidines adjacent to each other in the DNA, and high-energy ionizing radiation, which can cause double-strand breaks). Bases are also altered or lost spontaneously from mammalian DNA at a rate of many thousands per cell per day. If the damage is not repaired, a permanent change (mutation) is introduced that can result in any of a number of deleterious effects, including loss of control over the proliferation of the mutated cell, leading to cancer. Luckily, cells are remarkably efficient at repairing damage done to their DNA. Most of the repair systems involve recognition of the damage (lesion) on the DNA, removal or excision of the damage, replacement or filling the gap left by excision using the sister strand as a template for DNA synthesis, and ligation. These excision repair systems remove one to tens of nucleotides. [Note: Repair synthesis of DNA can occur outside of the S phase.]
A. Repair of mismatched bases (mismatch repair)
Sometimes replication
errors escape the proofreading function during DNA synthesis, causing a
mismatch of one to several bases. In E. coli, mismatch repair (MMR) is mediated
by a group of proteins known as the Mut proteins (Figure 29.28). Homologous
proteins are present in humans. [Note: MMR reduces the error rate of
replication from one in ten million to one in a billion.]
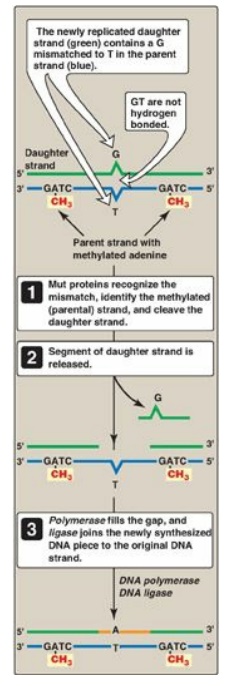
Figure 29.28 Methyl-directed mismatch repair in E. coli. [Note: The Mut proteins, S and L, recognize the mismatch and identify the parental (methylated) strand, and Mut H cleaves the daughter strand.] A = adenine; C= cytosine; G = guanine; T = thymine.
1. Identification of the mismatched strand: When a mismatch occurs, the Mut
proteins that identify the mispaired nucleotide(s) must be able to discriminate
between the correct strand and the strand with the mismatch. Discrimination is
based on the degree of methylation. GATC sequences, which are found
approximately once every thousand nucleotides, are methylated on the adenine
(A) residue. This methylation is not done immediately after synthesis, so the
newly synthesized DNA is hemimethylated (that is, the parental strand is
methylated, but the daughter strand is not). The methylated parental strand is
assumed to be correct, and it is the daughter strand that gets repaired. [Note:
The exact mechanism by which the daughter strand is identified in eukaryotes is
not yet known.]
2. Repair of damaged DNA: When the strand containing the
mismatch is identified, an endonuclease nicks the strand, and the mismatched
nucleotide(s) is/are removed by an exonuclease. Additional nucleotides at the 5I - and 3I -ends of the mismatch are also removed. The gap left by removal
of the nucleotides is filled, using the sister strand as a template, by a DNA
polymerase, typically DNA pol I. The 3I
-hydroxyl of the newly synthesized DNA is joined to the 5I -phosphate of the remaining stretch of the original DNA strand by
DNA ligase.
Mutation to the proteins involved in mismatch
repair in humans is associated with hereditary nonpolyposis colorectal cancer
(HNPCC), also known as Lynch syndrome. With HNPCC, there is an increased risk
for developing colon cancer (as well as other cancers); however, only about 5%
of all colon cancer is the result of mutations in mismatch repair.
B. Repair of damage caused by ultraviolet light (nucleotide excision repair)
Exposure of a cell to
ultrtaviolet (UV) light can result in the covalent joining of two adjacent
pyrimidines (usually thymines), producing a dimer. These thymine dimers prevent
DNA pol from replicating the DNA strand beyond the site of dimer formation.
Thymine dimers are excised in bacteria by UvrABC proteins in a process known as
nucleotide excision repair (NER) as illustrated in Figure 29.29. A related
pathway is present in humans (see below).
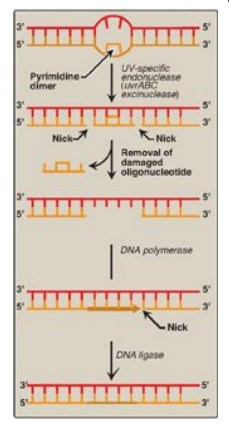
Figure 29.29 Nucleotide excision repair of pyrimidine dimers in E. coli DNA. UV = ultraviolet.
1. Recognition and excision of dimers by
UV-specific endonuclease:
First, a UV-specific endonuclease (called uvrABC excinuclease) recognizes the
dimer and cleaves the damaged strand on both the 5I -side and 3I -side of
the dimer. A short oligonucleotide containing the dimer is released, leaving a
gap in the DNA strand that formerly contained the dimer. This gap is filled in
using a DNA polymerase and DNA ligase.
2. UV radiation and cancer: Pyrimidine dimers can be formed in the skin cells of humans exposed to unfiltered sunlight. In the rare genetic disease xeroderma pigmentosum (XP), the cells cannot repair the damaged DNA, resulting in extensive accumulation of mutations and, consequently, early and numerous skin cancers (Figure 29.30). XP can be caused by defects in any of the several genes that code for the XP proteins required for nucleotide excision repair of UV damage in humans.
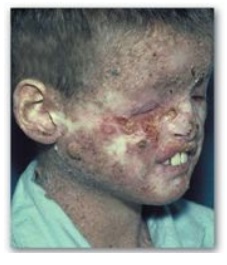
Figure 29.30 Patient with xeroderma pigmentosum.
C. Repair of base alterations (base excision repair)
The bases of DNA can be altered, either spontaneously, as is the case with cytosine, which slowly undergoes deamination (the loss of its amino group) to form uracil, or by the action of deaminating or alkylating compounds. For example, nitrous acid, which is formed by the cell from precursors such as the nitrosamines, nitrites, and nitrates, is a potent compound that deaminates cytosine, adenine (to hypoxanthine), and guanine (to xanthine). Bases can also be lost spontaneously. For example, approximately 10,000 purine bases are lost this way per cell per day. Lesions involving base alterations or loss can be corrected by base excision repair ([BER] Figure 29.31).
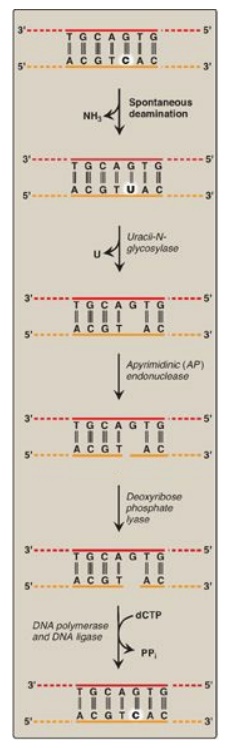
Figure 29.31 Correction of base alterations by base excision repair. T= thymine; A = adenine; C = cytosine; G = guanine; PPi = pyrophosphate.
1. Removal of abnormal bases: In BER, abnormal bases, such as
uracil, which can occur in DNA either by deamination of cytosine or improper
use of dUTP instead of dTTP during DNA synthesis, are recognized by specific
glycosylases that hydrolytically cleave them from the deoxyribose–phosphate
backbone of the strand. This leaves an apyrimidinic site (or apurinic, if a
purine was removed), both referred to as AP sites.
2. Recognition and repair of an AP site: Specific AP-endonucleases recognize that a base is missing and initiate the process of excision and gap-filling by making an endonucleolytic cut just to the 5I -side of the AP site. A deoxyribose phosphate lyase removes the single, base-free, sugar phosphate residue. DNA polymerase and DNA ligase complete the repair process.
D. Repair of double-strand breaks
Ionizing radiation or oxidative free radicals can cause double-strand breaks in DNA that are potentially lethal to the cell. Such breaks also occur naturally during gene rearrangements. dsDNA breaks cannot be corrected by the previously described strategy of excising the damage on one strand and using the remaining strand as a template for replacing the missing nucleotide(s). Instead, they are repaired by one of two systems. The first is nonhomologous end-joining (NHEJ), in which a group of proteins mediates the recognition, processing, and ligation of the ends of two DNA fragments. However, some DNA is lost in the process. Consequently, this mechanism of repair is error prone and mutagenic. Defects in this repair system are associated with a predisposition to cancer and immunodeficiency syndromes. The second repair system, homologous recombination (HR), uses the enzymes that normally perform genetic recombination between homologous chromosomes during meiosis. This system is much less error prone than NHEJ because any DNA that was lost is replaced using homologous DNA as a template. [Note: Mutations to the proteins, BRCA1 or 2 (breast cancer 1 or 2), which are involved in HR, increase the risk for developing breast cancer.]
Related Topics
