Classification of Microbes or Microorganisms
| Home | | Pharmaceutical Microbiology | | Pharmaceutical Microbiology |Chapter: Pharmaceutical Microbiology : Characterization, Classification and Taxonomy of Microbes
After having determined and established the characteristic variants of the microorganisms and documented methodically, the important task of their classification may be initiated and accomplished ultimately.
CLASSIFICATION
After
having determined and established the characteristic variants of the
microorganisms and documented methodically, the important task of their
classification may be initiated and accomplished ultimately.
1. Difficulties Encountered in Classification of Microorganisms
A large
cross section of microorganisms are found to be haploid* in nature, and they invariably undergo reproduction by asexual methods. Perhaps
that could be the most appropriate logical explana-tion that the concepts of
the species, as it is widely applicable to the plant and animal kingdoms that
normally reproduce sexually and wherein the species may be stated precisely
either in genetic or in evolutionary terms, can never be made
applicable very intimately and strictly to the microorganisms in the
right prespective. Importantly, the microbial species reasoning correctly can
never be regarded as an ‘interbreeding
population’ ; and, therefore, the two ensuing offspring caused by the
ultimate division of a microbial
cell are virtually quite ‘free’ to
develop in an altogether divergent fashion. It has been duly observed that the
reduction in genetic isolation caused by following two recombination procedures,
namely:
(a) Sexual
or para sexual recombination, and
(b) Special
mechanisms of recombination.
usually
offer great difficulty in assessing accurately the genuine effect of these
recombination phenom-ena by virtue of the fact that in nature the prevailing
frequencies with which they take place remain to be established. Nevertheless,
in the domain of microorganisms, the problem of reduction in ‘genetic isola-tion’ gets complicated by
the legitimate presence of the extrachromosomal** elements that specifi-cally help in the
chromosomal rearrangements and transfers as well.
In the
recent past, systematic and articulated attempts have been affected to
characterize the microbial species by carrying out the exhaustive descriptive
studies of both phenotype*** and geno-type****. Keeping in view the
remarkable simplicity as observed in the structural variants in the
micro-organisms these criteria or characteristics could not be used for their
systematic classification on a sound basis; and, therefore, one may resort to
alternative characteristic features, namely: genetic, biochemical,
physiological, and ecological aspects in order to supplement the structural
data authentically. Thus, one may infer conclusively that the bacterial classification is exclusively
employed as a supporting evi-dence more predominantly upon the functional attributes in comparison to
the structural attributes.
2. Objectives of Classification
Importantly,
the researchers and scientists practising ‘taxonomy’
i.e., the laws and principles of
classification of living organisms, do make great efforts to bring into being
logical and justifiable clas-sifications of microorganisms that essentially
possess the following two cardinal
qualities, namely:
(a) Stability : It has been duly observed that
such ‘classifications’ that are
essentially liable to experience rapid, radical alterations, practically
tantamount to utter confusion. Hence, sincere and ear nest efforts must be
geared into action to put forward such universally acceptable classifications
that would hardly require any major changes, whatsoever, as and when new
streams of information(s) crop up.
(b) Predictability: It is
ardently vital and important that by acquiring enough knowledge with respect to
the critical characteristic features of one specific bonafide member of a ‘taxonomic group’, it must be quite
possible and feasible to solemnly predict that the other members of the same
identical group presumably have almost similar characteristics as well. In
case, the said objective is not accom-plished satisfactorily, the ‘classification’ could be considered as
either invalid or of little value.
3. Genetic Methods of Classifying Microbes
There are
three most prominent ‘genetic methods’ that are invariably
employed for the methodi-cal arrangement of microbes based upon various
taxonomic groups (i.e., Taxa),
namely:
(i) Genetic
relatedness
(ii) The
intuitive method, and
(iii) Numerical
taxonomy.
The
aforesaid ‘genetic methods’ shall now
be treated separately in the sections that follows.
3.1. Genetic Relatedness
It is
regarded to be one of the most trustworthy and dependable method of
classification based solely upon the critical extent of genetic relatedness occurring between different organisms. In
addition this particular method is considered not only to be the utmost
objective of all other techniques based upon the greatest extent pertaining to
the fundamental aspect of organisms, but also their inherent he-reditary
material (deoxyribonucleic acid, DNA).
It is,
however, pertinent to state here that in actual practice the genetic relatedness may also be
estimated by precisely measuring the degree of hybridization taking place either between denatured DNA molecules
or between single stranded DNA and RNA species. The extent of homology* is as-sayed by strategically
mixing two different, types of ‘single-stranded DNA’ or ‘single-stranded DNA with RNA’ under highly specific and
suitable experimental parameters; and subsequently, measuring accurately the degree to which they
are actually and intimately associated to give rise to the formation of the
desired ‘double-stranded structures’
ultimately. The aforesaid aims and objectives may be accom-plished most
precisely and conveniently by rendering either the DNA or RNA radioactive and
measur-ing the radio activities by the help of Scintillation Counter or Geiger-Müller
Counter.
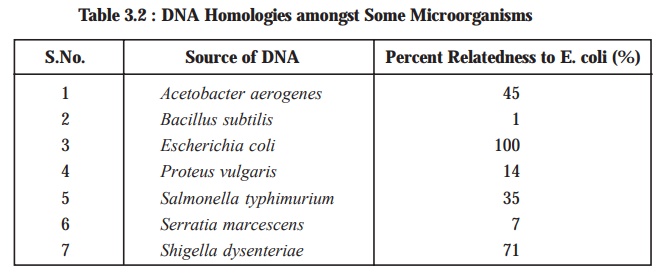
Table
3.2, shows the extent of genetic
relatedness of different microbes as assayed by the ensu-ing DNA-RNA hybridization. Nevertheless, it
has been duly demonstrated and proved that the genetic relatedness can be
estimated accurately by DNA-RNA
hybridization; however, the DNA-DNA
hybridi-zation affords the most precise results, provided adequate
precautions are duly taken to ascertain and
ensure that the prevailing hybridization
between the two strands is perfectly uniform.
3.2. The Intuitive Method
Various ‘microbiologists’ who have acquired
enormous strength of knowledge, wisdom, and hands-on experience in the
expanding field of ‘microbiology’
may at a particular material time vehe-mently decide and pronounce their
ultimate verdict whether the microorganisms represent one or more species or
genera. The most predominant and utterly important disadvantage of this particular method being that the
characteristic features of an organism which may appear to be critical and
vital to one researcher may not seem to be important to the same extent to
another, and altogether different taxono-mists would ultimately decide on
something quite different categorization at the end. Nevertheless, there are
certain ‘classification schemes’
that are exclusively based upon the intuitive
method and definitively proved to be immensely beneficial and useful in microbiology.
3.3. Numerical Taxonomy
The
survey of literatures have amply proved that in the Nineteenth Century, microbes were categorically grouped
strictly in proportion to their evolutionary affinities. Consequently, the
systematic and methodical segregation and arrangement of microorganisms into
the various organized groups was entirely on the specialized foundation of
inherited and stable structural and physiological characteristic features. This
arrangement is termed as the ‘Natural
Classification’ or the ‘Phylogenetic
Classifiction’.
Interestingly,
this particular modus operandi for
the classification of microorganisms has now almost turned out to be absolutely
redundant, and hence abandoned outright quite in favour of a rather more
realistic empirical approach based exclusively on ‘precise quantification’ pertaining to close similarities and distinct
dissimilarities prevailing amongst the various microbes. Michael Adanson was the first and
foremost microbiologist who unequivocally suggested this magnanimous approach,
which was termed as Adansonian Taxonomy
or Numerical Taxonomy.
Salient Features: The
various salient features of the
Numerical Taxonomy (or Adansonian
Taxonomy) are as enumerated below:
(1) The
fundamental basis of Numerical Taxonomy
is the critical assumption, that in the event when each phenotypic character is
assigned even and equal weightage, it must be viable and feasible to express
numerically the explicit taxonomic
distances existing between microor-ganisms, with regard to the number of actual characters which are shared in
comparison to the total number of
characters being examined ultimately. The importance of the Numerical Taxonomy is largely
influenced by the number of characters being investigated. Therefore, it would be absolutely necessary to
accomplish precisely an extremely high degree of signifi-cance—one should
examine an equally large number of characters.
(2) Similarity Coefficient and Matching
Coefficient: The determination of the similarity co efficient as well as the matching coefficient of any two
microbial strains, as characterized with regard to several character
variants viz., a, b, c, d etc., may
be determined as stated under:
Number of
characters + ve in both strains = a
Number of
characters + ve in ‘strain-1’ and – ve in ‘Strain-2’ = b
Number of
characters, – ve in ‘Strain-1’ and + ve in ‘Strain-2’ = c
Number of
characters – ve in both strain = d
Similarity
coefficient [Sj] = a / ( a + b + c )
Matching
coefficient [Ss] = a + b / ( a + b + c + d )
Based on
the results obtained from different experimental designs, it has been observed
that the similarity coefficient does
not take into consideration the characters that are ‘negative’ for both organ-isms; whereas, the matching coefficient essentially includes both positive and
negative characters.
Similarity Matrix: The ‘data’ thus generated are carefully
arranged in a ‘similarity matrix’ only
after having estimated the similarity
coefficient and the matching
coefficient for almost all microor-ganisms under investigation duly and
pair-wise, as depicted in Fig. 3.1 below. Subsequently, all these matrices may
be systematically recorded to bring together the identical and similar strains
very much close to one another.
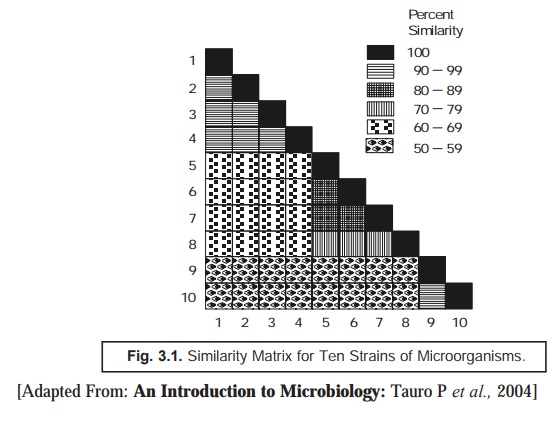
In actual
practice, such data are duly incorporated and transposed to a ‘dandogram’* as illus-trated in Fig.
3.2 under, that forms the fundamental basis for establishing the most probable
taxonomic
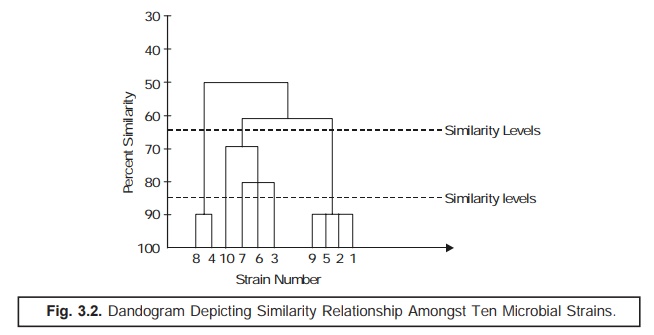
The ‘dotted line’ as indicated in
(Fig. 3.2) a dandogram evidently shows ‘similarity
levels’ that might be intimately
taken into consideration for recognizing
two different taxonomic ranks, for instance: a genus and a species.
The ‘Numerical Taxonomy’ or ‘Adansonian Approach’ was thought and
believed to be quite impractical and cumbersome in actual operation on account
of the reasonably copious volume and mag-nitude of the ensuing numerical
calculations involved directly. Importantly, this particular aspect has now
almost been eliminated completely by the advent of most sophisticated ‘computers’ that may be programmed
appropriately for the computation of the data, and ultimately, arrive at the degree of simi-larity with great ease,
simplicity, and precision. It is, however, pertinent to point out at this
juncture that though the ensuing ‘Numerical Taxonomy’ fails to throw any
light with specific reference to the pre-vailing genetic relationship, yet it
amply gives rise to a fairly stable fundamental basis for the articu-lated categorization of the
taxonomic distribution and groupings.
Limitations of Numerical Taxonomy: The
various limitations of numerical
taxonomy are as enumerated
under:
(1) It is
useful to classify strains within a larger group which usually shares the
prominent characteristic features in common.
(2) The
conventional classification of organisms solely depends on the observations and
knowledge of the individual taxonomist in particular to determine the ensuing
matching similarities existing between the bacterial strains; whereas, numerical taxonomy exclusively depends
upon the mathematical figures plotted on paper.
(3) The
actual usage of several tests reveals a good number of phenotypes, thereby more
genes are being screened; and, therefore, no organism shall ever be missed in
doing so.
(4) One
major limitation of the numerical
analysis is that in some instances, a specific strain may be grouped with a
group of strains in accordance to the majority of identical characteristic
features, but certainly not to all the prevailing characters. However,
simultaneously the particu-lar strain may possess a very low ebb of similarity
with certain other members of the cluster.
(5) The
exact location of the taxon is not
yet decided, and hence cannot be grouped or related to any particular taxonomic
group, for instance : genes or species.
(6) Evidently,
in the numerical analysis, the
definition of a species is not
acceptable as yet, whereas some surveys do ascertain that a 65% single-linkage cluster distincly
provides a 75% approximate idea of the specific species.
4. Systematized Classification
After
having studied the various aspects of characterization of microbes followed by the preliminary discussions on certain
important features related to their classification, one may now have an
ex-plicit broader vision on the systematized
classification. An extensive and intensive survey of literature would
reveal that the microorganisms may be classified in a systematized manner under
the following eight categories,
namely:
(i) Natural
classification,
(ii) Phyletic
classification,
(iii) Linnean
binomial scheme,
(iv) Phenotypic
classification,
(v) Microscopic
examination,
(vi) Cataloguing
rRNA,
(vii) Computer-aided
classification, and
(viii) Bacterial
classification (Bergey’s Manual of Systematic Bacteriology).
The
aforesaid eight categories in the
systematized classification of microorganisms would now be dealt with
individually in the sections that follows.
4.1. Natural Classification
The natural classification may be
considered as one of the most desirable classification systems which is broadly
based upon the anatomical characteristic features of the specific
microorganisms. In actual practice, the natural
classification predominantly helps to organize and arrange the wide
spec-trum of organisms into various categories (or groups) whose members do
share several characteristics, and reflects to the greatest extent the
intricate and complex biological nature of organisms. In reality, a plethora of
taxonomists have concertedly opined that a larger segment of the so called natural classifi-cation is importantly
and essentially the one having the maximum informations incorporated into it or the emanated predicted values obtained
thereof.
4.2. Phyletic* Classification
Phyletic classification usually
refers to the evolutionary development of
a species. Based upon the most
spectacular and master piece publication of Darwin’s—On the Origin of Species (1859), microbiologists across the globe
started making an attempt much to sincere and vigorous, so as to develop phyletic (or phylogenetic) classification
systems. Interestingly, the present system serves exclusively as a
supporting evidence on the evolutionary relationships in comparison to the
general resemblance. It has offered an appreciable hindrance for bacteria and
other microorganisms basically on account of the paucity of reliable and
authentic fossil records.
Nevertheless, the availability of most recent up to date copious volumes of
genuine information(s) with reference to comparison of genetic material and gene products, for instance: DNA, RNA,
proteins etc., mostly circumvent and overcome a large segment of these problems invariably encountered.
4.3. Linnean Binomial Scheme
The
microorganisms are invariably classified according to the Linnean Binomial Scheme of various genus and species. The International Code of Nomenclature of
Bacteria (ICNB) particularly specifies the scientific nomenclature (names)
of all categories (taxa) solely based
upon the following guidelines, namely:
(1) The ‘words’ used to refer to any taxonomic
group are either to be drawn from Latin
or are Latinized, if taken from
other languages.
(2) Each
distinct species is assigned a name comprising of two words viz., Salmonella
typhi; Bacillus subtilis ; and
the like. Here, the first word is the
name of the genus and is always written with a capital letter, whereas the second
word is a particular epithet (i.e., a descrip-tive word) which is not capitalized at all.
(3) A
taxonomic sequence of taxonomic groups is usually employed to categorize the
intimately related microorganisms at different stages of similarity. These categories or taxa are enu-merated as under:
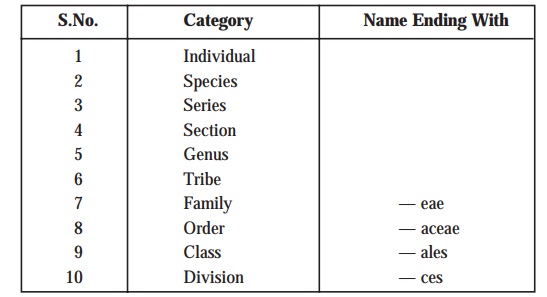
Explanations: The terminologies, species or genus are invariably employed as in the case of other types of classification. A species may be defined as a single type
of bacterium, whereas a genus
essentially includes a cluster of species all of which predominantly possess
substantial resemblance to one another to be considered intimately related;
and, therefore, may be distinguished very conveniently from the respective
bonafide members of the other genera. Importantly, the boundaries of certain gen-era are defined explicitly and
sharply; whereas, the boundaries of
species are relatively difficult and
cumbersome to define precisely.
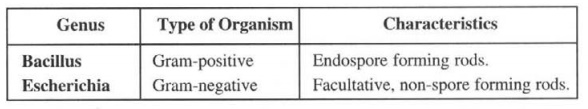
Example: The genus Bacillus can be evidently distinguished from the genus Escherichia as follows:
There are
three terminologies that are used very commonly in ‘microbiology’ e.g., strain,
clone, and a type species, which may be further expatiated as follows :
Strain: A stock, say of bacteria or
protozoa from a specific source and maintained in successive cultures or animal
inoculation.
Clone: It refers to the asexual progeny
of a single cell.
A Type Species: It is a culture that is
thoroughly studied and easily identifiable as a species. The ‘name’ of a type
species mostly conveys the prevailing characteristic features of the group.
4.4. Phenotypic Classification
The
spectacular and classical Adansonian approach of classifying microorganisms is
exclusively based upon the phenotypic characteristic features found in them. In
reality, such characteristics are overwhelmingly regarded as critical
expressions of a plethora of genes i.e., the basic unit of heredity made of
DNA, which essentially regulate and control the inherent cellular activities
via enzymes. Interestingly, it has been now universally accepted that the
phenotype ideally represents the reflection of the DNA base sequence.
Therefore, the best practicable and suitable methodology ot distinguishing two
individual organisms must be based upon the composition of their genetic
material. Quite recently sufficient advancement and substantial progress has
gained ground with regard to the genetic characterization of various
microorganisms, such as:
(a) analysis
of the base composition of DNA viz., to estimate the mole per cent of guanine
and cytosine in DNA (% G + C), and
(b) determination
of the extent of similarity existing between two DNA samples by causing
hybridization either between DNA & DNA or DNA & KNA. The fundamental
basis of this test is that the degree of hybridization would grossly serve as
an indication of the degree of relationship existing between the two DNA
samples (i.e., homology).
The DNA
of microbes significantly contains four bases : adenine (A), guanine (G),
thymine CD, and cytosine (C). and in a double-stranded DNA molecule usually, A
pairs with T and G pairs with C. However, the relative percentage of guanine
and cytosine may be expressed as follows:
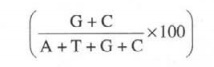
which
varies mostly with bacterial variants actually. Importantly, the composition of
chromosomal DNA deems to be fixed property of each cell which is distinctly
independent of age as well as other vital external influences.
Determination of % (G + C) of Chromosomal DNA: The
various steps involved in the determination of % (G + C) of chromosomal DNA are
as stated under:
(1) Extraction
of DNA from the cells by causing rupture very carefully and meticulously.
(2) The
resulting DNA is subject to purification to get rid of the non-chromosomal DNA.
(3) Subsequently,
the base composition may be estimated by adopting either of the following two methodologies, namely:
(a) Subjecting
the purified DNA to a gradually elevating temperature and determining the
ultimate enhancement in hypochromicity*,
and
(b) Centrifuging
the resulting DNA in cesium chloride
in density gradients.
Principle of Melting Point Method [i.e., Method 3(a)] : In an event when the double-stranded DNA is subject to enhancing
temperature, the two DNA strands undergo separation at a characteristic
temperature. The critical melting temperature solely depends on the actual (G +
C) content of the DNA. It has been duly observed that higher the (G + C)
content, higher shall be the melting point.
(4) Melting Point (Tm) : The particular mean
temperature at which the thermal denaturation of DNA takes
place is usually termed as the Melting
Point (Tm). However, Tm may be determined by
recording carefully the ‘observed change’
in the optical density of DNA
solution at 260 nm in the course of heating period, as illustrated in Fig. 3.3.
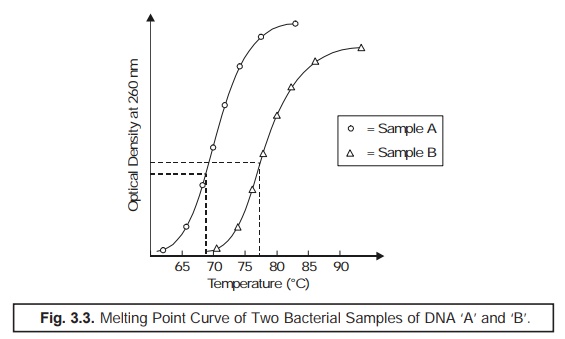
From the ‘melting point curve’ (Fig. 3.3) the
mole % (G + C) may be calculated by the help of the following expression:
%(G + C) = Tm × 63.54/0.47.
(5) Density Gradient Centrifugation: The % (G
+ C) composition may also be calculated by
estimating the relative rate of sedimentation in a cesium chloride
solution. In actual practice, the DNA preparations on being subjected to ultracentrifugation in the presence of a
heavy salt solution, shall emerge as a sediment at a specific region in
the centrifuge tube where its density
is equivalent to the density of the medium. Importantly, this method is
particularly suitable for such DNA samples that are heterogeneous in nature,
and hence could be sepa-rated simultaneously. It has been observed that the
ensuing buoyant density is an
extremely characteristic feature of each individual type of DNA; and hence is
solely dependent on the
(G + C)
values as shown in Fig. 3.4.
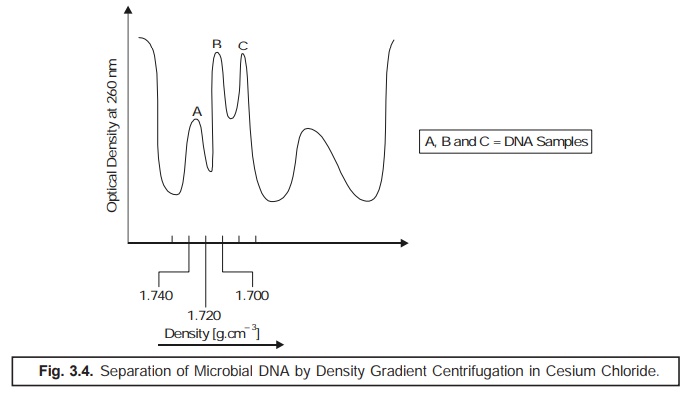
By the
help of buoyant density, it is quite
easy and convenient to arrive at the % (G + C) content precisely by employing
the following empirical formula:
P = 1.660
+ 0.00098 [% (G + C)] g . cm–3
(6) Chromatographic Method: Another
alternative method of estimating % (G + C) is accom-plished by the controlled hydrolysis of DNA in the
presence of acids, separating the nucleotides
by ultracentrifugation, and ultimately assaying the nucleotides by
chromatogra-phy. Though this method is apparently lengthy and tedious, yet is
quite simple and gives reasonably accurate results.
4.5. Microscopic Examination
In
general, microorganisms have been duly classified by microscopic examination based upon their shape, size, and various
staining characteristics. It has been abundantly proved that the stained preparations have obviously provided much better and clear
information ; however, the unstained
preparations may also be employed for these investigations to a certain
extent as well.
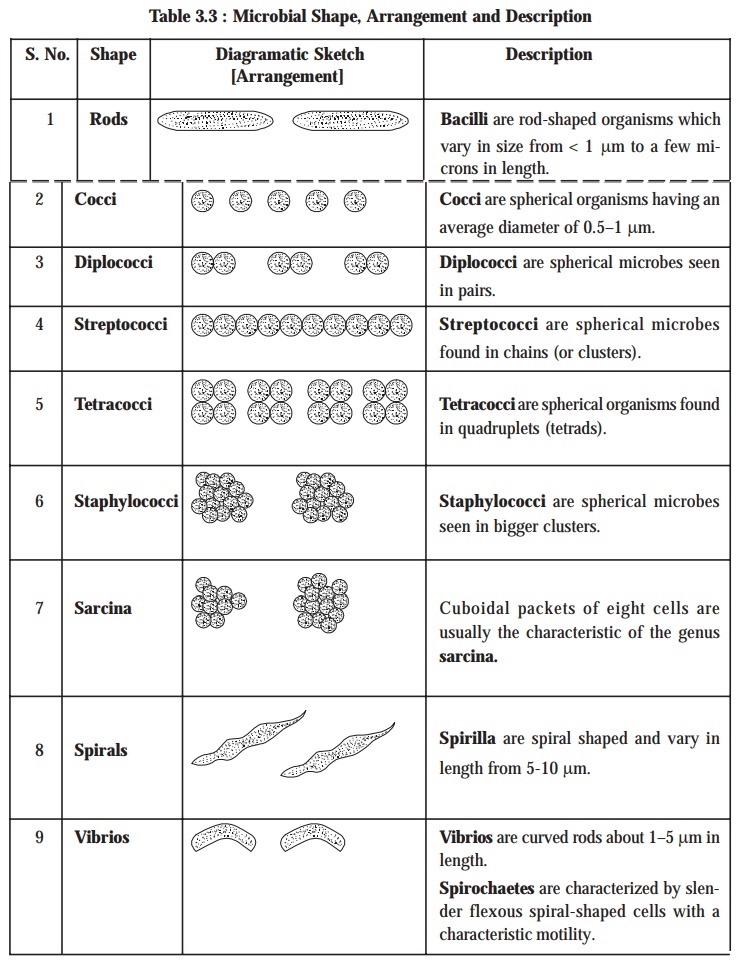
The size and shape of microbes invariably may provide sufficient valuable
informations that may be gainfully utilized for the presumptive diagnostic identification, as depicted in the following
Table 3.3:
4.6. Cataloguing rRNA
Since mid
seventies, progressive comparative
analysis of the 16 S rRNA sequences
had gained a tremendous momentum which enabled its proper and legitimate usage
to explore the prokaryotic phylogeny. The ribosomal RNA (i.e.,
rRNA) molecules are found to be of immense choice due to the following three cardinal reasons:
(a) They
exhibit a constant function,
(b) They
are universally present in all organisms, and
(c) They
seem to have changed in sequence extremely slowly.
Salient Features.
The
various salient features in cataloguing rRNA are as enumerated under:
(1) 5S rRNA
Molecule: Because of its relatively smaller size it has been taken as an accurate indicator of the phylogenetic relationship.
(2) 16S rRNA
Molecule: It is sufficiently large ; and, therefore, quite easy to handle with a reasonably high degree of precision.
(3) 23S rRNA
Molecule: Because of its relatively much larger size it is rather more difficult
to characterize, and hence used in
the comparative analysis.
(4) In
the last two decades, the 16 S rRNA
has been critically examined, explored, and extracted from a large
cross-section of microorganisms and duly digested with ribonuclease T1. The resulting nucleotide are
meticulously resolved by 2D-electrophoresis*
technique, and sequenced appropriately.
(5) The
advent of latest sophisticated instrument e.g.,
DNA-Probe** which may sequence
nucleic acids have further aided in the phenomenon of sequencing of 16 S rRNA from microorganisms.
(6) The
skilful comparison of rRNA catalogues
predominantly designates genealogical
rela-tionship existing amongst the wide range of microbes.
(7) The
aforesaid genealogical relationship
may be suitably quantified in terms of an associa-tion
coefficient, designated as SAB, which proves to be a typical
characteristic feature for a pair of
microorganisms. The association coefficient SAB may be expressed as follows :
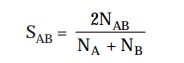
where, NAB
= Number of residues existing in sequences common to two rRNA catalogues.
NA
and NB = Total number of residues duly represented by oligomers of
at least 6 nucleotides in catalogues A and B respectively.
(8) As to
date, the rRNA sequences of more than
200 species of microbes and eukaryotes
have been duly characterized and documented adequately.
(9) It
has been observed that most of the microorganisms strategically give rise to a
coherent but also a very large segment including the eubacteria. Importantly, the methanogens, halophiles, and
thermoacidophiles do not necessarily fall within the domain of eubacteria***.
(10) The
aforesaid kind of rRNA sequencing has
in fact duly permitted the methodical and logi-cal characterization of archaeobacteria.
4.7. Computer Aided Classification
In the
latest spectacular and astronomical growth in the field of computer technology, it has inducted a tremendous impetus and great
help in the proper grouping of microorganisms, and eventually classifying them
with an utmost accuracy and precision. One may come across a host of problems
in comparing a relatively huge number of characteristic features as may be seen
in the very instance of numerical
taxonomy or the Adansonian approach under
the perview of the general classification of microbes. In order to circumvent such difficulties and problems,
the proper usage of computer-aided programmes and devices have been
rightly pressed into service for determining the differentiating capacity
of the tests and also for determining
the overall similarity with the known organisms. As to date, the commendable extremely high speed and memory of computer conveniently allows it to
accomodate very swiftly a host of possible species in the
identification/classification phenomenon by judiciously comparing the
characteristic properties of an ‘unknown
microorganism’ with those stored duly in the computer. In fact, the advent of the utility of computer, definitely and grossly
minimizes the probability of error in the identification/classification by
virtue of either infrequent occurrence of a microorganism or the critical
presence of a rather more frequent microbe with not-so similar or superfi-cial
resemblance to other organisms. A good number of highly sophisticated, modern,
and advanced computer softwares (systems)
for microbiology have now
been duly developed and put into practice
across the world profusely. The ‘microbiological
laboratories’ strategically attached to most modern hospitals and research and development (R & D)
laboratories have gainfully commenced the utiliza-tion of the elaborated computer facilities in the
handling/processing of ‘test samples’
to obtain most reliable, dependable, and reproducible results meant to be used
in correct diagnosis and research activi-ties with certainly more confidence
and fervour.
4.8. Bacterial Classification [Bergey’s Manual of Systematic Bacteriology]
Microorganisms represent an exceptionally large
conglomerate of minute living body with enor-mous diversity having a
procaryotic cellular organization. Several sincere intensive and extensive
studies were duly made with particular reference to their broad spectrum physical,
structural, and functional characteristic qualities, but none of them could
ever produce and evolve an overall satisfactory generally acceptable
classification.
Chester
(1899 and 1901) initiated and took active interest in the classification of
bacteria, and subsequently published for the first time—‘The Manual of Determinative Bacteriology’. The said manual was
painstakingly and meticulously revised, substantiated, and modified by David
Hendrick’s Bergey (1923) and entitled as—‘Bergey’s
Manual of Systematic Bacteriology’, later on commonly termed as ‘Bergey’s Manual’. In fact, Bergey’s Manual is being recognized as
the ‘official compen-dium of all
identified and classified bacteria, and serves as an indispensable and valuable
guide to the microbiologists across the globe.
The
latest edition of ‘Bergey’s Manual’—(1994)
provides a more rational and emperical ap-proach for the classification of
bacteria. Besides, it gives rise to an effective system of keys for
establish-ing the precise genetic
position of an unknown organism. Table 3.4 gives a comprehensive account of
the classification of bacteria (Division II)* upto the generic level.
Table 3.4. Summary of Bacterial Classification [Bergey’s Manual — 1994]
Part-1: Phototrophic Bacteria
Order I : Rhodospirillales
Suborder
I : Rhodospirillineae
Family I
: Rhodospirillaceae
Genus I :
Rhodospirillum
Genus II
: Rhodopseudomonas
Genus III
: Rhodopseudomonas
Family II
: Chromatiaceae
Genus I :
Chromatium
Genus II
: Thiocystis
Genus III
: Thiosarcina
Genus IV
: Thiospirilum
Genus V :
Thiocapsa
Genus VI
: Lamprocystis
Genus VII
: Thiodicatyon
Genus
VIII : Thiopedia
Genus IX
: Amoebobacter
Genus X :
Ectothiorhodospira
Suborder
: Chlorobineae
Family
III : Chlorobiaceae
Genus I :
Chlorobium
Genus II
: Prosthecocloris
Genus III
: Chloropseudomonas
Genus IV
: Pelodictyon
Genus V :
Clathrochloris
Incertae
Sedis [Addenda]
Genus :
Chlorochromatium
Genus :
Cylindrogloea
Genus :
Chlorobacterium
Part-2: Gliding Bacteria
Order I : Myxobacterales
Family I
: Myxococcaceae
Genus I :
Myxococcus
Family II
: Archangiaceae
Genus I :
Archangium
Family
III : Cystobacteraceae
Genus I : Cystobacter
Genus II
: Melittangium
Genus III
: Stigmatella
Family IV
: Polyangiaceae
Genus I :
Polyangium
Genus II
: Nannocystis
Genus III
: Chondromyces
Order II : Cytophagales
Family I
: Cytophagaceae
Genus I :
Cytophaga
Genus II
: Flexibacter
Genus III
: Herpetosiphon
Genus IV
: Flexibacter
Genus V :
Saprospira
Genus VI
: Sporocytophaga
Family II
: Beggiatoaceae
Genus I :
Beggiatoa
Genus II
: Vitreoscilla
Genus III
: Thioploea
Family
III : Simonsiellaceae
Genus I :
Simonsiella
Genus II
: Alysiella
Family IV
: Leucotrichaceae
Genus I :
Leucothrix
Genus II
: Thiothrix
Incertae
Sedis [Addenda]
Genus :
Toxothrix
Familiae
incertae sedis
Achromatiaceae
Genus :
Achromatium
Pelonemataceae
Genus I :
Pelonema
Genus II
: Achronema
Genus III
: Peloploca
Genus IV
: Desmanthos
Part-3: Sheathed Bacteria
Genus :
Sphaerotilus
Genus :
Leptothrix
Genus :
Streptothrix
Genus :
Lieskeela
Genus :
Phragmidiothrix
Genus :
Crenothrix
Genus :
Clonothrix
Part-4: Budding And/Or Appendaged Bacteria
Genus :
Hyphomicrobium
Genus :
Hyphomonas
Genus :
Pedomicrobium
Genus :
Caulobacter
Genus :
Asticeacaulis
Genus :
Ancalomicrobium
Genus :
Prosthecomicrobium
Genus :
Thiodendron
Genus :
Pasteuria
Genus :
Blastobacter
Genus :
Seliberia
Genus :
Gallionella
Genus :
Nevskia
Genus :
Planctomyces
Genus :
Metallogenium
Genus :
Caulococcus
Genus :
Kusnezonia
Part-5: Spirochaetes
Order I : Spirochaetales
Family I
: Spirochaetaceae
Genus I :
Spirochaeta
Genus II
: Cristispira
Genus III
: Treponema
Genus IV
: Borrelia
Genus V :
Leptospira
Part-6: Spiral And Curved Bacteria
Family I
: Spirillaceae
Genus I :
Spirillum
Genus II
: Campylobacter
Incertae
Sedis [Addenda]
Genus :
Bdellovibrio
Genus :
Microcyclus
Genus :
Pelosigma
Genus :
Brachyarcus
Part-7: Gram-Negative Aerobic Rods And Cocci
Family I
: Pseudomonadaceae
Genus I :
Pseudomonas
Genus II
: Xanthomonas
Genus III
: Zoogloea
Genus IV
: Gluconobacter
Family II
: Azotobacteraceae
Genus I :
Azotobacter
Genus II
: Azomonas
Genus III
: Beijerinckia
Genus IV
: Derxia
Family
III : Rhizobiaceae
Genus I :
Rhizobium
Genus II
: Agrobacterium
Family IV
: Methylomonadaceae
Genus I :
Methylomonas
Genus II
: Methylococcus
Family V
: Halobacteriaceae
Genus I :
Halobacterium
Genus II
: Halococcus
Incertae
Sedis [Addenda]
Genus :
Alcaligenes
Genus :
Acetobacter
Genus :
Brucella
Genus :
Bordetella
Genus :
Francisella
Genus :
Thermus
Part-8: Gram-Negative Facultatively Anaerobic Rods
Family I
: Enterobacteriaceae
Genus I :
Escherichia
Genus II
: Edwardsiella
Genus III
: Citrobacter
Genus IV
: Salmonella
Genus V :
Shigella
Genus VI
: Klebsiella
Genus VII
: Enterobacter
Genus
VIII : Hafnia
Genus IX
: Serratia
Genus X : Proteus
Genus XI : Yersinia
Genus XII : Erwinia
Family II : Vibrionaceae
Genus I : Vibrio
Genus II : Acromonas
Genus III : Plesiomonas
Genus IV : Photobacterium
Genus V : Lucibacterium
Incertae Sedis [Addenda]
Genus : Chromobacterium
Genus : Zymomonas
Genus : Flavobacterium
Genus : Haemophilus
Genus : Pasteurella
Genus : Actinobacillus
Genus : Cardiobacterium
Genus : Streptobacillus
Genus : Calymmatobacterium
Part-9: Gram-Negative Anaerobic Bacteria
Family I : Bacteriodaceae
Genus I : Bacteroides
Genus II : Fusobacterium
Genus III : Leptotrichia
Incertae Sedis [Addenda]
Genus : Desulfovibrio
Genus : Butyrivibrio
Genus : Succinivibrio
Genus : Succinimonas
Genus : Lachnospira
Genus : Selenomonas
Part-10: Gram-Negative Cocci And Coccobacilli
Family I : Neisseriaceae
Genus I : Neisseria
Genus II : Branhamella
Genus III : Moraxella
Genus IV : Acinetobacter
Incertae Sedis [Addenda]
Genus : Paracoccus
Genus : Lampropedia
Part-11: Gram-Negative Anaerobic Cocci
Family I : Veillonellaceae
Genus I : Acidaminococcus
Genus II : Veillonella
Genus III : Megasphaera
Part-12: Gram-Negative Chemolithotrophic Bacteria
Family 1 : Nitrobacteraceae
Genus I : Nitrobacter
Genus II : Nitrospina
Genus III : Nitrococcus
Genus IV : Nitrosomonas
Genus V : Nitrospira
Genus VI : Nitrosococcus
Genus VII : Nitrosolobus
Organisms Metabolizing Sulphur
Genus I : Thiobacillus
Genus II : Sulfolobus
Genus III : Thiobacterium
Genus IV : Macromonas
Genus V : Thiovulum
Genus VI : Thiospira
Family II : Siderocapsaceae
Genus I : Siderocapsa
Genus II : Naumanniella
Genus III : Ochrobium
Genus IV : Siderococcus
Part-13: Methane Producing Bacteria
Family I : Methanobacteriaceae
Genus I : Methanobacterium
Genus II : Methanosarcina
Genus III : Methanococcus
Part-14: Gram Positive Cocci
Family I : Micrococcaceae
Genus I : Micrococcus
Genus II : Staphylococcus
Genus III : Planococcus
Family II : Streptococcaceae
Genus I : Streptococcus
Genus II : Leuconostoc
Genus III : Pediococcus
Genus IV : Acrococcus
Genus V : Gemella
Family III : Peptococcaceae
Genus I : Peptococcus
Genus II : Peptostreptococcus
Genus III : Ruminococcus
Genus IV : Sarcina
Part-15: Endospore Forming Rods and Cocci
Family I : Bacillaceae
Genus I : Bacillus
Genus II : Sporolactobacillus
Genus III : Clostridium
Genus IV : Desulfotomaculum
Genus V : Sporosarcina
Incertae Sedis [Addenda]
Genus : Oscillospira
Part-16: Gram-Positive Asporogenous Rod-Shaped
Bacteria
Family I : Lactobacillaceae
Genus I : Lactobacillus
Incertae Sedis [Addenda]
Genus : Listeria
Genus : Erysipelothrix
Genus : Caryophanon
Part-17: Actinomycetes And Related Organisms
Coryneform
Group of Bacteria
Genus I : Corynebacterium
Genus II : Arthrobacter
Incertae Sedis [Addenda]
Genus A : Brevibacterium
Genus B : Microbacterium
Genus III : Cellulomonas
Genus IV : Kurthia
Family I : Propionibacteriaceae
Genus I : Propionibacterium
Genus II : Eubacterium
Order I : Actinomycetales
Family I : Actinomycetaceae
Genus I : Actinomyces
Genus II : Arachnia
Genus III : Bifidobacterium
Genus IV : Bacterionema
Genus V : Rothia
Family II : Mycobacteriaceae
Genus I : Mycobacterium
Family III : Frankiaceae
Genus I : Frankia
Family IV : Actinoplanaceae
Genus I : Actinoplanes
Genus II : Spirillospora
Genus III : Streptosporangium
Genus IV : Amorphosphorangium
Genus V : Ampullariella
Genus VI : Pilimelia
Genus VII : Planomonospora
Genus VIII : Planobispora
Genus IX : Dactylosporangium
Genus X : Kitastoa
Family V : Dermatophillaceae
Genus I : Dermatophilus
Genus II : Geodermatophilus
Family VI : Nocardiaceae
Genus I : Nocardia
Genus II : Pseudonocardia
Family VII : Streptomycetaceae
Genus I : Streptomyces
Genus II : Streptoverticilium
Genus III : Sporichthya
Genus IV : Microellobosporia
Family VIII : Micromonosporaceae
Genus I : Micromonospora
Genus II : Thermoactinomyces
Genus III : Actinobifida
Genus IV : Thermonospora
Genus V : Microbispora
Gebus VI : Micropolyspora
Part-18: Rickettsias
Order I : Rickettsiales
Family : Rickettsiaceae
Tribe I : Rickettsieae
Genus I : Rickettsia
Genus II
: Rochalimaea
Genus III
: Coxiella
Tribe II
: Ehrlichieae
Genus IV
: Ehrlichia
Genus V :
Cowdria
Genus VI
: Neorickettsia
Tribe III
: Wolbachieae
Genus VII
: Wolbachia
Genus
VIII : Symbiotes
Genus IX
: Blattabacterium
Genus X :
Rickettsiella
Family :
Bartonellaceae
Genus I :
Bartonella
Genus II
: Grahamella
Family :
Anaplasmataceae
Genus I :
Anaplasma
Genus II
: Paranaplasma
Genus III
: Aegyptionella
Genus IV
: Haemobartonella
Genus V :
Eperythrozoon
Order II
: Chlamydiales
Family I
: Chlamydiaceae
Genus I :
Chlamydia
Part-19 :
Mycoplasmas
Class
Mollicutes
Order I :
Mycoplasmatales
Family I
: Mycoplasmataceae
Genus I :
Mycoplasma
Family II
: Acholeplasmataceae
Genus I :
Acholeplasma
Incertae
Sedis [Addenda]
Genus :
Thermoplasma
Incertae
Sedis [Addenda]
Genus :
Spiroplasma
Related Topics
