The Changing Paradigm of Drug Development and Delivery
| Home | | Pharmacovigilance |Chapter: Pharmacovigilance: Pharmacogenetics and the Genetic Basis of ADRs
Some sequence polymorphisms that are involved in disease pathogenesis also may be involved in determining drug response, directly or indirectly. One example is apolipoprotein E4.
THE CHANGING PARADIGM OF DRUG
DEVELOPMENT AND DELIVERY
OTHER RELEVANT POLYMORPHISMS
Some
sequence polymorphisms that are involved in disease pathogenesis also may be
involved in determining drug response, directly or indirectly. One example is
apolipoprotein E4 (Apo-E4), a risk factor for familial late-onset and sporadic
Alzheimer’s Disease (AD) that has been reported to predict poor response to the
cholinesterase inhibitor tacrine (Farlow et
al., 1996). The presence of the E4 allele also may be a factor in the
success of prophylactic oestrogen therapy for AD (Sadee, 2000). In addi-tion,
Apo-E4 is associated with increased risk of coronary artery disease (CAD).
Gerdes et al. (2000) reported that
presence of this allele is associated with almost 2-fold increased risk of
death in myocardial infarct survivors and that this increased mortality rate
can be abolished by treatment with simvastatin
(HMG-CoA/3-hydroxy-3methylglutaryl coenzyme A reductase inhibitor). Increased
understanding of the underlying multigenic causes of AD, CAD and other diseases
and neurodegenerative disorders may lead to the development of strategies for
disease treatment and even prophylaxis for those at high risk of devel-oping a
disease.
Another
example of a disease-related polymor-phism that is predictive of drug response
involves cholesteryl ester transfer protein (CETP) and pravas-tatin (HMG-CoA
reductase inhibitor used to treat hypercholesterolemia). Kuivenhoven et al. (1998) reported a significant
relationship between variation of the CETP gene and the progression of
coro-nary atherosclerosis, independent of lipolytic plasma enzyme activity and
plasma HDL cholesterol concen-tration. If these results are replicated, then
the pres-ence of a homozygous polymorphism at this site could be used to
predict whether treatment with pravastatin will be effective. Replication of
the study has led to variable results, but Boekholdt
et al. (2005), conducted
a meta analysis of seven large
population-based studies (total patients >3500) and two randomized placebo
control trials. They found that Taq1B polymorphism was associated with HDL-C
and subsequent CAD, but not with Pravastatin therapy.
CURRENT CHALLENGES IN THE CLINICAL APPLICATION OF PHARMACOGENETIC KNOWLEDGE
It
is clear from this cursory review of the current state of knowledge regarding
the genetic basis of ADRs that many clinically significant genetic
poly-morphisms affecting drug response in humans have been described already.
The emphasis to date has been on identification of mutant alleles at a single
gene locus (e.g. Phase I and II hepatic enzymes, TPMT and DPyDH), and this
research has been fruitful. However, drug response depends on the drug’s
inter-action with the many proteins involved in its absorp-tion, distribution,
excretion and target site, each of which is coded for by genes that may be
associated with common variants that may affect response. For example, one
individual may exhibit polymorphisms of genes coding for two drug-related
proteins: one that affects the degree of drug inactivation and one that
determines the sensitivity of the drug receptor. The polymorphism affecting the
drug’s metabolism would determine the plasma concentrations to which the
individual is exposed, and the polymorphic recep-tor would determine the nature
of the individual’s response at a given drug concentration. These poly-genic
interactions are much more difficult to establish during the course of clinical
drug trials than are the monogenic effects discussed above but may have an even
more significant impact on drug response.
The
effects of environmental factors may be modi-fied by wild-type or polymorphic
genes, as well, intro-ducing more confounding variables. The majority of these
gene variants are relatively uncommon in the general population, making it
difficult to establish their role in drug response and demonstrate clini-cal
relevance, especially for heterozygous individuals who are likely to exhibit
more subtle effects than homozygotes.
Clinical
research in disease genetics and pharma-cogenetics has, at times, produced
discordant and contradictory results, creating confusion and result-ing in a
lack of credibility in the minds of many health care providers. Inconsistent
results may be due to several factors, including the lack of strict diagnostic
criteria for study entry, the heterogeneous nature of the diseases being
studied, the use of differ-ent end points and scales for assessing drug
efficacy and ADRS, the presence of unknown or unidenti-fied environmental
factors and the polygenic nature of many drug effects (Evans and Relling, 1999).
It is crucial in clinical pharmacogenetic research that the study sample be of
adequate size to demonstrate the necessary statistical power and that the
results be rigorously confirmed in comparable populations by other researchers
(Manasco, Rieser and Pericak-Vance, 2000). In addition, once a drug has been
approved, ongoing, systematic centralized collection of meaningful, evaluable
data regarding drug efficacy and ADRs does not occur routinely, and
pharmacoge-netic data are rarely collected at all. As a result, oppor-tunities
for increasing our knowledge of dramatic and subtle genetic effects on drug
response both in large numbers of diverse patients and specific diagnostic
subsets of patients are lost.
Doroshow,
in his review of the gefitinib clinical trial and regulators at the FDA in
their guidance documents on drug/device co-development highlight the need to
prospectively collect biological samples linked to clinical data and consent
during Phase III trials (www.fda.gov/genomics). Experience from the development
of Herceptin, Iressa and now Tarceva highlight the need to have samples that
accompany the clinical data to enable market-ready tests to be developed,
reviewed and marketed at the time of drug release.
In
2004, Merck removed its COX2 inhibitor Vioxx from the marketplace after it was
found to be associ-ated with increased risk of cardiovascular side effects.
Considerable backlash directed towards the FDA and the pharmaceutical industry
resulted. The increased scrutiny provided the impetus for enhanced
pharma-covigilance efforts. Furthermore, there was a recog-nition that the AERS
voluntary reporting system was not adequate to fully evaluate post-marketing
safety.
Several
of the guidance documents highlighted the opportunity the pharmacogenomics can play
in iden-tifying populations at risk.
Much
pharmacogenetic research to date has involved identifying and categorizing
drug-related polymorphisms while relatively little has been done to determine
clinical relevance in well-defined populations. Clinicians do not know which
variants should be assessed, how and by whom that should be done, what drugs
might be affected, what course of action would be appropriate based on the
informa-tion obtained and who will cover the cost of the test. Should the dose
be altered? By how much? Should the drug be avoided entirely? What about
related drugs and polymorphisms? Must each be tested sepa-rately? What effects
do the variants have on drug–drug and drug–environment interactions? What
issues exist around professional liability and the ethical, legal and social
aspects of such testing? Carefully designed, well-controlled clinical studies
in appropriate popu-lations must be carried out to begin to answer these
pressing questions, and the information then must be made available to
clinicians and reflected in ethically grounded standards of clinical practice
and compen-sation procedures.
In
addition, standard pharmacotherapy references and treatment guidelines
formulated by various health care organizations rarely contain relevant
pharmaco-genetic information even when it is known, making it difficult for
clinicians to gain access to existing data. Consumers, health care providers,
payers and regu-latory agencies lack basic education with regard to
pharmacogenetics and timely access to relevant new data as they emerge.
Although the lay press occa-sionally spot-lights a tragedy that could have been
averted through the application of existing pharmaco-genetic knowledge (such as
‘overdose’ deaths of slow drug metabolizers; Stipp, 2000), the need for
increased professional and public awareness and education in this arena is
equal to the need for continuing research.
Until
recently, genotyping an individual was a labo-rious, time-consuming and costly
proposition that was undertaken only if there was a high index of suspi-cion of
an identified genetic disorder. The Human Genome Project and the multitude of
high technology spin-offs from it are changing this situation. Auto-mated
instrumentation, new bioinformatics systems and novel strategies derived from
genomic research will enable researchers to evaluate and analyze the wealth of
genetic information that will continue to emerge. High-throughput DNA
sequencing, gene mapping and transcriptional analyses are becoming economically
and scientifically feasible as a result of innovations such as DNA, cDNA
(‘edited’ version of a gene, containing only the parts that will be expressed
as proteins) and oligonucleotide microarrays and microfluidic analytical
devices (Mancinelli, Cronin and Sadee, 2000).
Because
genetic information does not change, it is conceivable that a consumer could
have their genetic sample or genetic information in a central location that is
available to healthcare providers in any location and at any time. The need for
point of care genetic tests will ultimately be eliminated.
In
contrast, tests that measure expression of a gene vary over time and thus the
need for dynamic measurement for gene expression or protein expres-sion will be
needed in some cases.
SINGLE NUCLEOTIDE POLYMORPHISMS, MEDICINE RESPONSE TESTS AND ‘GENETIC TESTS’
Many
of the previously mentioned polymorphisms directly alter the metabolism,
transport, action or excretion of medicines through identified (or
identifiable) structural or functional effects; there is a causal relationship
between the polymorphism and the phenotype. New genomic techniques such as
those mentioned above are making it possible to detect associations (which may or may not be causal) between specific
genetic markers and indi-vidual response to medicines. Single nucleotide
poly-morphisms (SNPs, pronounced ‘snips’), single-base differences in DNA
sequence, are the most common form of human polymorphisms. They occur with an
average frequency of about 1 per 1300 base pairs, serving as easily
identifiable virtual mileposts along the three billion base pair human genome
(Interna-tional Human Genome Sequencing Consortium, 2001) (See Figure 51.1).
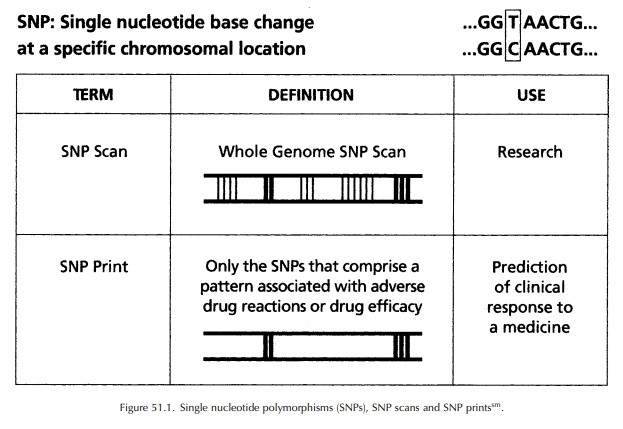
The
SNP Consortium (a not-for-profit organization of pharmaceutical and
bioinformational companies, academic centres and a charitable trust) produced
an ordered high-density SNP map of the human genome, which is publicly
available at http://snp.cshl.org. This map is being used to find disease genes
and to correlate genetic information with individual responses to medicines.
Although few of these SNPs are expected to be involved directly with disease or
medicine response, they will be useful as analytical tools to track small
segments of the genome. Indi-viduals who carry a particular gene variant
(allele) are likely to carry variants of several SNP markers that are close to
or within that allele because of the phenomenon of linkage disequilbrium (LD;
when alle-les are in close physical proximity, they are likely to be inherited
together).
The
results of comprehensive analysis of genetic variability in genes related to
the action of the medicine in question or the disease process as well as
whole-genome SNP scanning obtained during Phase II clinical trials of a
medicine can be used to iden-tify specific SNP markers, patterns or haplotypes
that correlate to patient responses (efficacy and ADRs). The medicine
response-related data could form the basis for the selection of patients most
likely to respond well in Phase III trials, possibly making these trials
smaller, faster and more efficient if the goal of the study is to validate
markers for efficacy. Studies designed to validate safety markers will require
larger numbers of patients due to the rate of the adverse events.
The
FDA has issued guidance related to co-development of a drug and predictive
test. This guid-ance is focused on development of a predictive test if the
markers are known at the time of Phase II trials. Unfortunately, this is not the
usual case for drug development, but the emphasis on how samples are collected
(with appropriate documented consent using processes that enable evaluation of
chain of custody) and the need for a prospective approach to studying and
replicating genetic findings will apply in other situations as well
(www.fda.gov/genomics).
The
FDA has also formed an interdisciplinary review group to review voluntary
genomic data submissions (VGDS). The VGDS process enables sponsors to submit
data to the agency and get feed-back without being required to submit the data
to the NDA. If genomic data are to be included as part of the label or as part
of the decision making process, then the VGDS process does not apply.
Functional
enzyme analysis of TPMT in red blood cells before treatment with specific
cancer chemother-apy drugs is one example of an MRT in current use in the
United States. Another is HercepTest®, which uses a polyclonal
antibody to detect HER2 protein, reflecting HER2 expression in breast cancer
cells; it is used to predict patient response to trastuzumab (Herceptin®),
a humanized monoclonal antibody against the HER2 receptor. Researchers already
have developed other tools for assessing HER2 expression, including a
monoclonal antibody test for the HER2 protein, a test for circulating HER2
protein (in the extracellular domain) and a test using fluorescence in situ hybridization (FISH) directly to
determine the number of copies of the HER2 gene (not its protein product).
Because of the strong corre-lation between overexpression of HER2 protein and
response to Herceptin®, the US FDA required that a test kit to
assess HER2 protein expression be commer-cially available before drug approval
– an example of a regulatory agency mandating the availability of a predictive
test linked to use of a specific drug.
Transcriptional
analyses, in which the expres-sion levels of DNA are measured, may provide
another approach for predictive tests for medicine response (Kleyn and Vesell,
1998). RNA obtained from biopsied tissue and surgical specimens can be used for
expression-based studies in some cancers, allowing detection of somatic changes
associated with the development of some tumours and their response to
chemotherapy. For example, the ampli-fication of the oncogene erb-B2 predicts a good response to
treatment with a specific adjuvant ther-apy
(cyclophosphamide-methotrexate-5-fluorouracil) for breast cancer (Muss et al., 1994). Alternately, the
expression of genes predicting drug response can be assayed at the protein
level using antibody-based tests of serum or other tissues (Kleyn and Vesell,
2000).
Ongoing
genetic and genomic research undoubt-edly will result in the development of
additional tools that can be incorporated into or used as the basis of medicine
response tests. The rationale exists for conducting pharmacogenetic analyses to
look for associations between drug responses (safety and effi-cacy) and
genotype, and the technology exists for conducting these analyses. The missing
piece is a pool of DNA samples with associated medical and medicine response
data to facilitate the efficient conduct of pharmacogenetic research.
Eventually, MRTs based on SNPs and other genetic polymor-phisms will enable
health care providers to identify patients at high risk of developing a given
disease, implement preventive therapy and lifestyle adjust-ments when
appropriate and choose the medicines that are most likely to benefit the
patient and least likely to result in serious ADRs (Mancinelli, Cronin and
Sadee, 2000).
In the past, the term ‘genetic testing’ has been asso-ciated with the diagnosis of monogenic diseases such as cystic fibrosis and Huntingdon disease – conditions for which a causative, single, genetic mutation has been identified (Table 51.4). A newer area of genetic research and testing involves identification of genes related to the occurrence of common complex diseases such as asthma, heart disease and migraine. These diseases are likely to result from the interaction of multiple ‘increased risk’ or susceptibility genes with each other and possibly with environmental factors. These types of genetic research and testing (related to monogenic diseases and susceptibility genes) involve determining the likelihood of occurrence (prediction) or the presence (diagnosis) of disease in individuals. Although very useful and important, there are social and ethical risks related to the nature of the infor-mation revealed by these tests – what it means, who has access to it and how it can be used. There is general agreement on the need for genetic counselling to help patients and families understand and process the results of these disease- and risk-related genetic tests.
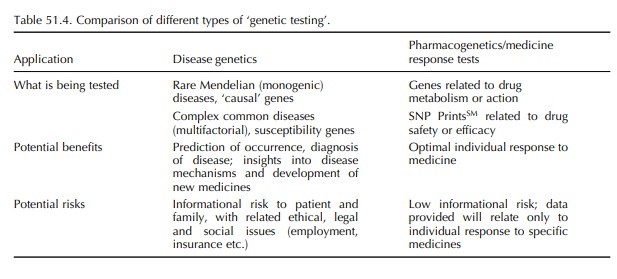
In
contrast, the risks associated with tests to detect polymorphisms related to
response to medicines, such as metabolic and drug receptor or target character
istics and genomic profiles, are minimal: the data that are obtained are quite
limited and specific to the drug(s) being considered. No information about
disease, causal or susceptibility genes, is likely to be obtained. These tests,
when validated, will be simi-lar to routine laboratory tests such as blood
typing, drug concentration monitoring and liver enzyme anal-yses. Although
health care providers would discuss the results with patients, there would be
no need for genetic counselling and ongoing psychosocial support related to
interpretation of the results, with rare exceptions.
Related Topics
