Steps in Protein Synthesis
| Home | | Biochemistry |Chapter: Biochemistry : Protein Synthesis
The process of protein synthesis translates the 3-letter alphabet of nucleotide sequences on mRNA into the 20-letter alphabet of amino acids that constitute proteins.
STEPS IN PROTEIN SYNTHESIS
The process of protein
synthesis translates the 3-letter alphabet of nucleotide sequences on mRNA into
the 20-letter alphabet of amino acids that constitute proteins. The mRNA is
translated from its 5I -end to its 3I -end, producing a protein synthesized
from its amino (N)-terminal end to its carboxyl (C)-terminal end. Prokaryotic
mRNAs often have several coding regions (that is, they are polycistronic;).
Each coding region has its own initiation and termination codon and produces a
separate species of polypeptide. In contrast, each eukaryotic mRNA has only one
coding region (that is, it is monocistronic). The process of translation is
divided into three separate steps: initiation, elongation, and termination.
Eukaryotic protein synthesis resembles that of prokaryotes in most aspects.
Individual differences are noted in the text.
One important difference is that translation and transcription are temporally linked in prokaryotes, with translation starting before transcription is completed as a consequence of the lack of a nuclear membrane in prokaryotes.
A. Initiation
Initiation of protein synthesis involves the assembly of the components of the translation system before peptide bond formation occurs. These components include the two ribosomal subunits, the mRNA to be translated, the aminoacyl-tRNA specified by the first codon in the message, GTP (which provides energy for the process), and initiation factors that facilitate the assembly of this initiation complex (see Figure 31.13). [Note: In prokaryotes, three initiation factors are known (IF-1, IF-2, and IF-3), whereas in eukaryotes, there are many (designated eIF to indicate eukaryotic origin). Eukaryotes also require ATP for initiation.] The following are two mechanisms by which the ribosome recognizes the nucleotide sequence (AUG) that initiates translation.
1. Shine-Dalgarno sequence: In Escherichia coli (E. coli), a
purine-rich sequence of nucleotide bases, known as the Shine-Dalgarno (SD)
sequence, is located six to ten bases upstream of the initiating AUG codon on
the mRNA molecule (that is, near its 5I -end). The 16S rRNA component of the
small (30S) ribosomal subunit has a nucleotide sequence near its 3-end that is
complementary to all or part of the SD sequence. Therefore, the 5-end of the
mRNA and the 3-end of the 16S rRNA can form complementary base pairs,
facilitating the positioning of the small ribosomal subunit on the mRNA in
close proximity to the initiating AUG codon (Figure 31.10).
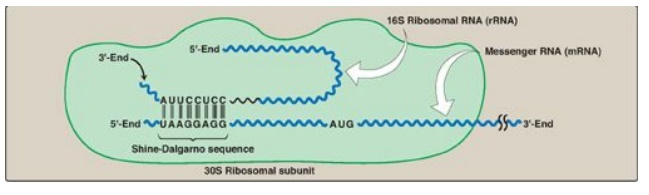
Figure 31.10 Complementary binding
between prokaryotic mRNA Shine-Dalgarno sequence and 16S rRNA. S = Svedberg
unit.
2. 5I Cap: Eukaryotic mRNAs do not have SD sequences. In
eukaryotes, the small (40S) ribosomal subunit (aided by members of the elF-4
family of proteins) binds close to the cap structure at the 5-end of the mRNA
and moves down the mRNA until it encounters the initiator AUG. This “scanning”
process requires ATP. [Note: Interactions between the cap-binding eIF-4
proteins and the poly-A tail-binding proteins on eukaryotic mRNA mediate
circularization of the mRNA and likely prevent the use of incompletely
processed mRNA in translation.]
3. Initiation codon: The initiating AUG is recognized
by a special initiator tRNA. Recognition is facilitated by IF-2-GTP in
prokaryotes and eIF-2-GTP (plus additional eIFs) in eukaryotes. The charged
initiator tRNA enters the P site on the small subunit. The initiator tRNA is
the only tRNA recognized by (e)IF-2 and the only tRNA to go directly to the P
site. In bacteria and in mitochondria, the initiator tRNA carries an
N-formylated methionine (fMet, Figure 31.11). After Met is attached to the initiator
tRNA, the formyl group is added by the enzyme transformylase, which uses N10-formyl
tetrahydrofolate as the carbon donor. In eukaryotes, the initiator tRNA carries
a Met that is not formylated. In both prokaryotic and eukaryotic cells, this
N-terminal Met is usually removed before translation is completed. The large
ribosomal subunit then joins the complex, and a functional ribosome is formed
with the charged initiating tRNA in the P site. The A site is empty. [Note:
Specific (e)IFs function as anti-association factors and prevent premature
addition of the large subunit.] The GTP on (e)IF-2 gets hydrolyzed to GDP. A
guanine nucleotide exchange factor facilitates the reactivation of (e)IF-2-GDP
through replacement of GDP by GTP.
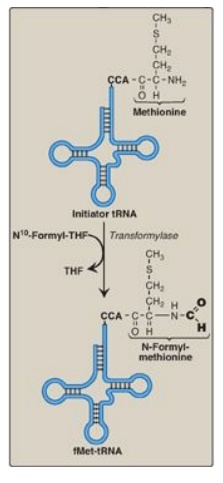
Figure 31.11 Generation of the initiator N-formylmethionyl-tRNA (fMet-tRNA). THF = tetrahydrofolate; C = cytosine; A = adenine.
B. Elongation
Elongation of the
polypeptide chain involves the addition of amino acids to the carboxyl end of
the growing chain. During elongation, the ribosome moves from the 5I - end to
the 3I -end of the mRNA that is being translated. Delivery of the
aminoacyl-tRNA whose codon appears next on the mRNA template in the ribosomal A
site (a process known as decoding) is facilitated in E. coli by elongation
factors EF-Tu-GTP and EF-Ts and requires GTP hydrolysis. [Note: In eukaryotes,
comparable elongation factors are EF-1a-GTP and EF-1bg. Both EF-Ts and EF-1bg
function in guanine nucleotide exchange.] The formation of the peptide bond is
catalyzed by peptidyltransferase, an activity intrinsic to the 23S rRNA found
in the large (50S) ribosomal subunit (Figure 31.12). [Note: Because this rRNA
catalyzes the reaction, it is referred to as a ribozyme.] After the peptide
bond has been formed, what was attached to the tRNA at the P site is now linked
to the amino acid on the tRNA at the A site. The ribosome then advances three
nucleotides toward the 3I -end of the mRNA. This process is known as
translocation and, in prokaryotes, requires the participation of EF-G-GTP
(eukaryotic cells use EF-2-GTP) and GTP hydrolysis. Translocation causes
movement of the uncharged tRNA from the P to the E site for release and
movement of the peptidyl-tRNA from the A to the P site. The process is repeated
until a termination codon is encountered.
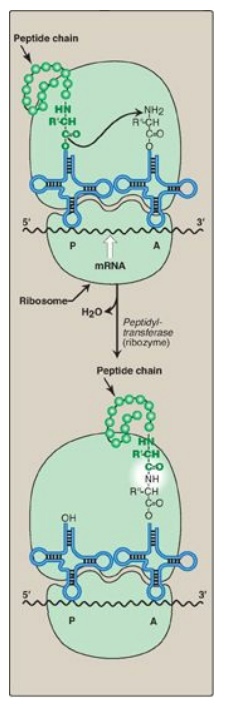
Figure 31.12 Formation of a peptide bond. Peptide bond formation involves transfer of the peptide on the transfer RNA (tRNA) in the P site to the amino acid on the tRNA in the
C. Termination
Termination occurs when
one of the three termination codons moves into the A site. These codons are
recognized in E. coli by release factors: RF-1, which recognizes the
termination codons UAA and UAG, and RF-2, which recognizes UGA and UAA. The
binding of these release factors results in hydrolysis of the bond linking the
peptide to the tRNA at the P site, causing the nascent protein to be released
from the ribosome. A third release factor, RF-3-GTP then causes the release of
RF-1 or RF-2 as GTP is hydrolyzed (see Figure 31.13). [Note: Eukaryotes have a
single release factor, eRF, which recognizes all three termination codons. A
second factor, eRF-3, functions like the prokaryotic RF-3. See Figure 31.15 for
a summary of the factors used in translation.] The steps in prokaryotic protein
synthesis are summarized in Figure 31.13. The newly synthesized polypeptide may
undergo further modification as described below, and the ribosomal subunits,
mRNA, tRNA, and protein factors can be recycled and used to synthesize another
polypeptide. [Note: In prokaryotes, ribosome recycling factors mediate
separation of the subunits.] Some antibiotic inhibitors of protein synthesis
are illustrated in Figure 31.13, as is diphtheria toxin.
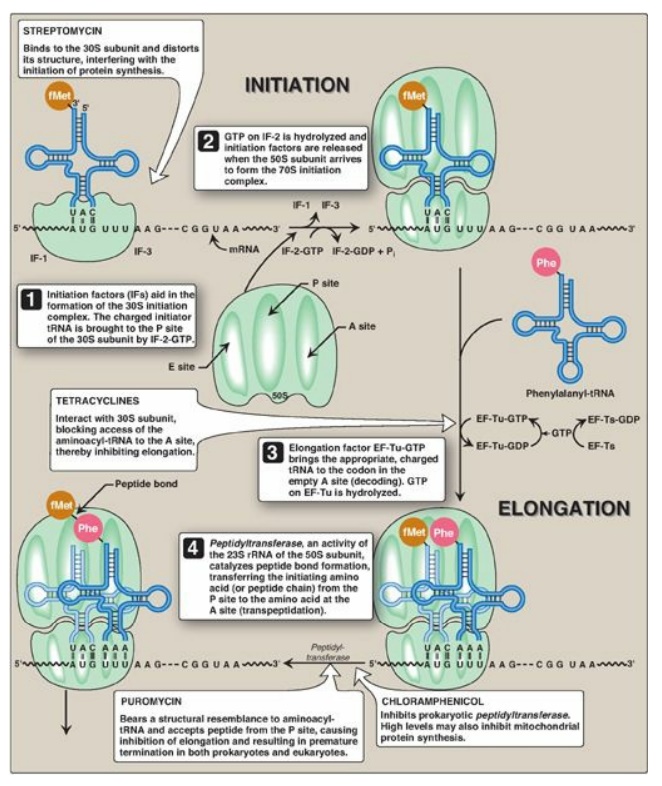
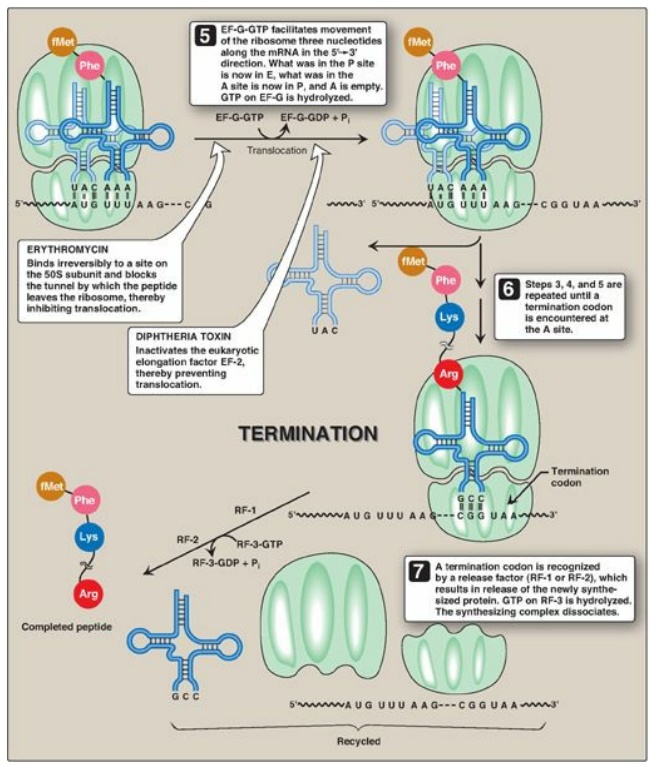
Figure 31.13 Steps in prokaryotic protein synthesis (translation), and their inhibition by antibiotics. [Note: EF-Ts is a guanine nucleotide exchange factor. It facilitates the removal of GDP, allowing its replacement by GTP. The eukaryotic equivalent is EF-1βγ.] fMet = formylated methionine; S = Svedberg unit; GTP = guanine nucleoside triphosphate; Phe = phenylalanine.
D. Polysomes
Translation begins at
the 5I -end of the mRNA, with the ribosome proceeding along the RNA molecule.
Because of the length of most mRNAs, more than one ribosome at a time can
translate a message (Figure 31.14). Such a complex of one mRNA and a number of
ribosomes is called a polysome or polyribosome.
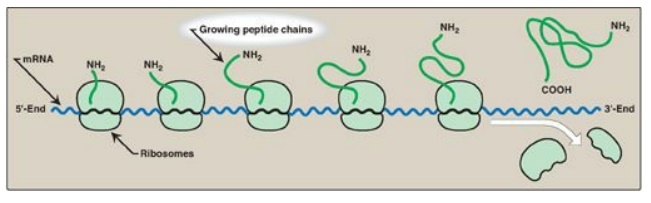
Figure 31.14 A polyribosome consists of several ribosomes simultaneously translating one messenger RNA (mRNA). [Note: Eukaryotic mRNA is circularized for translation.]
E. Regulation of translation
Gene expression is most
commonly regulated at the transcriptional level, but translation may also be
regulated. An important mechanism by which this is achieved in eukaryotes is by
covalent modification of eIF-2: phosphorylated eIF-2 is inactive. In both
eukaryotes and prokaryotes, regulation can also be achieved through proteins
that bind mRNA and inhibit its use by blocking translation or extend its use by
protecting it from degradation. For a more detailed discussion of the
regulation of translation.
F. Protein targeting
Although most protein synthesis in eukaryotes is initiated in the cytoplasm, many proteins perform their functions within subcellular organelles or outside of the cell. Such proteins usually contain amino acid sequences that direct the proteins to their final locations. For example, proteins destined for secretion from the cell are targeted during their synthesis (cotranslational targeting) to the RER by the presence of an N-terminal hydrophobic signal sequence. [Note: The sequence is recognized and bound by the signal recognition particle, which facilitates transport to the RER.] Proteins targeted after synthesis (posttranslational) include nuclear proteins that contain an internal, short, basic “nuclear localization signal” and mitochondrial matrix proteins that contain an N-terminal, amphipathic, α-helical “mitochondrial entry sequence.”
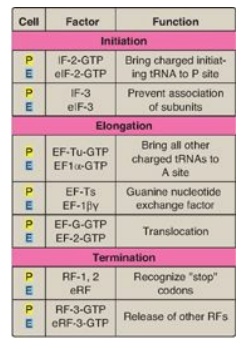
Figure 31.15 Protein factors
in the three stages of translation. P = prokaryotes;E = eukaryotes;
tRNA = transfer RNA; IF = initiation factor; EF = elongation factor; RF =
release factor.
Related Topics
