Organization of Eukaryotic DNA
| Home | | Biochemistry |Chapter: Biochemistry : DNA Structure, Replication, and Repair
A typical (diploid) human cell contains 46 chromosomes, whose total DNA is approximately 2 m long! It is difficult to imagine how such a large amount of genetic material can be effectively packaged into a volume the size of a cell nucleus so that it can be efficiently replicated, and its genetic information expressed.
ORGANIZATION OF EUKARYOTIC DNA
A typical (diploid)
human cell contains 46 chromosomes, whose total DNA is approximately 2 m long!
It is difficult to imagine how such a large amount of genetic material can be
effectively packaged into a volume the size of a cell nucleus so that it can be
efficiently replicated, and its genetic information expressed. To do so
requires the interaction of DNA with a large number of proteins, each of which
performs a specific function in the ordered packaging of these long molecules
of DNA. Eukaryotic DNA is associated with tightly bound basic proteins, called
histones. These serve to order the DNA into fundamental structural units,
called nucleosomes, which resemble beads on a string. Nucleosomes are further
arranged into increasingly more complex structures that organize and condense
the long DNA molecules into chromosomes that can be segregated during cell
division. [Note: The complex of DNA and protein found inside the nuclei of
eukaryotic cells is called chromatin.]
A. Histones and the formation of nucleosomes
There are five classes
of histones, designated H1, H2A, H2B, H3, and H4. These small, evolutionally
conserved proteins are positively charged at physiologic pH as a result of
their high content of lysine and arginine. Because of their positive charge,
they form ionic bonds with negatively charged DNA. Histones, along with ions
such as Mg2+, help neutralize the negatively charged DNA phosphate groups.
1. Nucleosomes: Two molecules each of H2A, H2B, H3, and H4 form the
octameric core of the individual nucleosome “beads.” Around this structural
core, a segment of dsDNA is wound nearly twice, causing supercoiling (Figure
29.26). [Note: The N-terminal ends of these histones can be acetylated,
methylated, or phosphorylated. These reversible covalent modifications
influence how tightly the histones bind to the DNA, thereby affecting the
expression of specific genes. Histone modification is an example of “epigenetics”
or heritable changes in gene expression without alteration of the nucleotide
sequence.] Neighboring nucleosomes are joined by “linker” DNA approximately 50
base pairs long. H1, of which there are several related species, is not found
in the nucleosome core, but instead binds to the linker DNA chain between the
nucleosome beads. H1 is the most tissue specific and species specific of the
histones. It facilitates the packing of nucleosomes into more compact
structures.
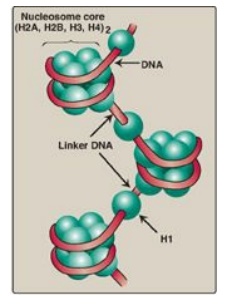
Figure 29.26 Organization of human DNA, illustrating the structure of nucleosomes. H = histone.
2. Higher levels of organization: Nucleosomes can be packed more
tightly to form a polynucleosome (also called a nucleofilament). This structure
assumes the shape of a coil, often referred to as a 30-nm fiber. The fiber is
organized into loops that are anchored by a nuclear scaffold containing several
proteins. Additional levels of organization lead to the final chromosomal
structure (Figure 29.27).
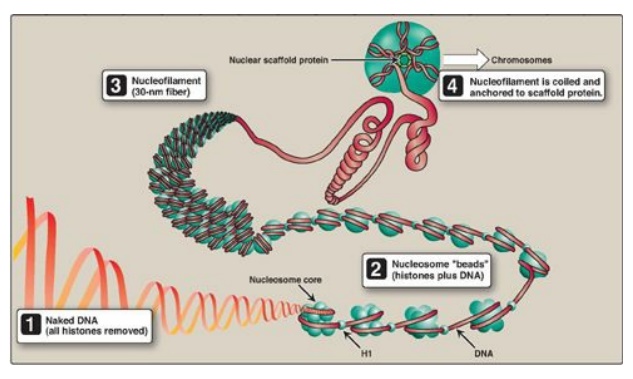
Figure 29.27 Structural organization of eukaryotic DNA. [Note: A 104 compaction is seen from to1 to 4 .] H = histone.
B. Fate of nucleosomes during DNA replication
Parental nucleosomes
are disassembled to allow access to DNA during replication. Once DNA is
synthesized, nucleosomes form rapidly. Their histone proteins come both from
new synthesis and from the transfer of intact parental histone octamers.
Related Topics
