Diagnosis of Infectious Diseases
| Home | | Pharmaceutical Microbiology | | Pharmaceutical Microbiology |Chapter: Pharmaceutical Microbiology : Recombinant DNA Technology
Each pathogenic microorganism contains genetic material that distinguishes it from its host and from other microorganisms. This specific material constitutes a signature that allows the identification of a particular microorganism from a complex mixed population.
DIAGNOSIS OF INFECTIOUS DISEASES
Each pathogenic
microorganism contains genetic material that distinguishes it from its host and from other microorganisms. This specific material
constitutes a signature that allows
the identification of a particular microorganism from a complex
mixed population. In the diagnosis of infectious diseases, identification of specific sequences from microbial pathogens
will allow appropriate treatment at an early stage
as well as prevention of the
spread of disease.
The two
main techniques used
for the diagnosis of infectious disease
targeting nucleic acids
are hybridization and PCR-based
amplification. There are currently
specific primers
and probes for
the detection of more
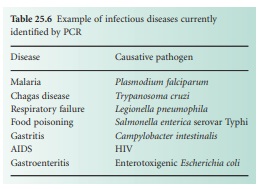
than 100 infectious disease. Table 25.6 shows just
a few examples.
A) DNA Hybridization Techniques
Nucleic acid
hybridization is based
on the precise nucleotide base pairing and hydrogen bonding
between one strand of nucleotides and a complementary nucleotide sequence. Any diagnostic
nucleic acid hybridization consists of three essential elements:
a DNA probe, the target DNA and the signal
detection system. Recent developments
in detection systems
and improvements in safety have enabled
the use of highly sensitive nonradioactive
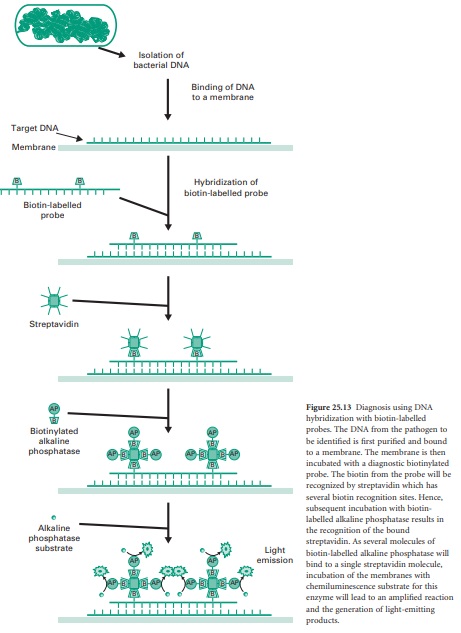
methods. Figure 25.13 illustrates the general steps required for DNA hybridization using chemiluminescent-based detection. This
non-radioactive system achieves signal
amplification by enzymatic conversion of a chemiluminescent substrate. First of all, DNA from the biological sample including the target DNA from the pathogen to be identified needs to be extracted.
A diagnostic biotin-labelled probe is then mixed
with the target DNA bound
to a membrane support. The
hybridized probe is then incubated with streptavidin, which
has multiple sites that avidly bind
biotin. Subsequent incubation with biotin
conjugated to alkaline
phosphatase results in the enzymatic labelling of the bound streptavidin. Finally,
addition of a special substrate for the alkaline phosphatase
results in the conversion of this substrate into a product which emits light and which can be detected after exposure
to X-ray film or by using sensitive imaging cameras.
The procedure described above can
also be scaled down, automated and redesigned to use thousands
of sequence-specific probes
at once in what are called microarrays. In this case non-labelled DNA probes are synthesized and minute
droplets are spotted
at high density
on to glass slides
where they will remain bound.
The DNA from the biological sample
is then labelled at the extremities with a fluorescent dye and after denaturation into single strands it is allowed
to anneal with
the probes bound on the slide. After
washing, labelled DNA fragments which annealed
to specific probes are visualized by illuminating the slide with light of the appropriate wavelength to cause the dyes to fluoresce. As microarrays can carry thousands of probes per square centimetre, this is performed by automated
scanning devices and dedicated
software which
identifies the probes
producing positive signals. This technique allows
the screening at once of a
very large number of
pathogens or defined genetic markers.
B) PCR Amplification Using Fluorescent Primers
As in many other
fields, PCR has brought a revolutionary
change in nucleic acid-based diagnosis. In a clinical setting, PCR has many desirable features such as the sensitivity to work
with tiny amounts
of DNA samples from blood or tissue
to achieve a specific and significant amplification of target sequences. Furthermore, the rapidity
of this process, as explained previously in this chapter, provides a significant advantage in the
early diagnosis and treatment of infectious diseases.
A PCR fluorescence-based technique
has been used successfully
in the diagnosis of infectious diseases. In this case the PCR primers are
labelled with a fluorescent dye that is bound
to the 5′ end of each primer.
Two main types of fluorescent dyes are normally used: one is fluorescein, which
appears green under
certain light wavelengths, and the other
is rhodamine, which
appears red. After PCR amplification of the target
sequence with the fluorescent-labelled primers, the primers
are removed by chromatographic separation, and the presence of
the labelled PCR product is detected. The absence of labelled
PCR product
is interpreted as the absence
of the target DNA sequence.
The procedure
can also be applied to detect specific RNAs, for example to detect RNA viruses. In this case
the extracted RNA has to be converted first
to ds cDNA with reverse transcriptase before the PCR
amplification. The procedure
is then called reverse transcriptase PCR or
RT-PCR.
Related Topics
